5 Oracle R Connector for Hadoop
This chapter describes R support for big data. It contains the following sections:
5.1 About Oracle R Connector for Hadoop
Oracle R Connector for Hadoop is a collection of R packages that provide:
-
Interfaces to work with Hive tables, the Apache Hadoop compute infrastructure, the local R environment, and Oracle database tables
-
Predictive analytic techniques, written in R or Java as Hadoop MapReduce jobs, that can be applied to data in HDFS files
You install and load this package as you would any other R package. Using simple R functions, you can perform tasks like these:
-
Access and transform HDFS data using a Hive-enabled transparency layer
-
Use the R language for writing mappers and reducers
-
Copy data between R memory, the local file system, HDFS, Hive, and Oracle databases
-
Schedule R programs to execute as Hadoop MapReduce jobs and return the results to any of those locations
Several analytic algorithms are available in Oracle R Connector for Hadoop: linear regression, neural networks for prediction, matrix completion using low rank matrix factorization, clustering, and non-negative matrix factorization. They are written in either Java or R.
To use Oracle R Connector for Hadoop, you should be familiar with MapReduce programming, R programming, and statistical methods.
Oracle R Connector for Hadoop APIs
Oracle R Connector for Hadoop provides access from a local R client to Apache Hadoop using functions with these prefixes:
-
hadoop: Identifies functions that provide an interface to Hadoop MapReduce -
hdfs: Identifies functions that provide an interface to HDFS -
orch: Identifies a variety of functions;orchis a general prefix forORCHfunctions -
ore: Identifies functions that provide an interface to a Hive data store
Oracle R Connector for Hadoop uses data frames as the primary object type, but it can also operate on vectors and matrices to exchange data with HDFS. The APIs support the numeric, integer, and character data types in R.
All of the APIs are included in the ORCH library. The functions are listed in Chapter 6 in alphabetical order.
See Also:
The R Project website athttp://www.r-project.org/5.2 Access to HDFS Files
For Oracle R Connector for Hadoop to access the data stored in HDFS, the input files must comply with the following requirements:
-
All input files for a MapReduce job must be stored in one directory as the parts of one logical file. Any valid HDFS directory name and file name extensions are acceptable.
-
Any file in that directory with a name beginning with an underscore (_) is ignored.
-
The input files must be in comma-separated value (CSV) format as follows:
-
key := string | ""
-
value := string[,value]
-
line := [key\t]value
-
string := char[string]
-
char := regexp([^,\n])
-
The following are examples of acceptable CSV input files:
-
CSV files with a key:
"1\tHello,world" "2\tHi,there"
-
CSV files with no key:
"Hello,world" "Hi,there"
-
CSV files with a NULL key:
"\tHello,world" "\tHi,there"
5.3 Access to Hive
Hive provides an alternative storage and retrieval mechanism to HDFS files through a querying language called HiveQL, which closely resembles SQL. Hive uses MapReduce for distributed processing. However, the data is structured and has additional metadata to support data discovery. Oracle R Connector for Hadoop uses the data preparation and analysis features of HiveQL, while enabling you to use R language constructs.
See Also:
The Apache Hive website athttp://hive.apache.org5.3.1 ORE Functions for Hive
You can connect to Hive and manage objects using R functions that have an ore prefix, such as ore.connect. If you are also using Oracle R Enterprise, then you recognize these functions. The ore functions in Oracle R Enterprise create and manage objects in an Oracle database, and the ore functions in Oracle R Connector for Hadoop create and manage objects in a Hive database. You can connect to one database at a time, either Hive or Oracle Database, but not both simultaneously.
The following ORE functions are supported in Oracle R Connector for Hadoop:
as.ore as.ore.character as.ore.frame as.ore.integer as.ore.logical as.ore.numeric as.ore.vector is.ore is.ore.character is.ore.frame is.ore.integer is.ore.logical is.ore.numeric is.ore.vector ore.create ore.drop ore.get ore.pull ore.push ore.recode
This release does not support ore.factor, ore.list, or ore.object.
These aggregate functions from OREStats are also supported:
aggregate fivenum IQR median quantile sd var*
*For vectors only
5.3.2 Generic R Functions Supported in Hive
Oracle R Connector for Hadoop also overloads the following standard generic R functions with methods to work with Hive objects.
- Character methods
-
casefold,chartr,gsub,nchar,substr,substring,tolower,toupperThis release does not support
greplorsub. - Frame methods
-
-
attach,show -
[,$,$<-,[[,[[<- -
Subset functions:
head,tail -
Metadata functions:
dim,length,NROW,nrow,NCOL,ncol,names,names<-,colnames,colnames<- -
Conversion functions:
as.data.frame,as.env,as.list -
Arithmetic operators:
+,-,*,^,%%,%/%,/ -
Compare,Logic,xor,! -
Test functions:
is.finite,is.infinite,is.na,is.nan -
Mathematical transformations:
abs,acos,asin,atan,ceiling,cos,exp,expm1,floor,log,log10,log1p,log2,logb,round,sign,sin,sqrt,tan,trunc -
Basic statistics:
colMeans,colSums,rowMeans,rowSums,Summary,summary,unique -
by,merge -
unlist,rbind,cbind,data.frame,eval
This release does not support
dimnames,interaction,max.col,row.names,row.names<-,scale,split,subset,transform,with, orwithin. -
- Logical methods
-
ifelse,Logic,xor,! - Matrix methods
-
Not supported
- Numeric methods
-
-
Arithmetic operators:
+,-,*,^,%%,%/%,/ -
Test functions:
is.finite,is.infinite,is.nan -
abs,acos,asin,atan,ceiling,cos,exp,expm1,floor,log,log1p,log2,log10,logb,mean,round,sign,sin,sqrt,Summary,summary,tan,trunc,zapsmall
This release does not support
atan2,besselI,besselK,besselJ,besselY,diff,factorial,lfactorial,pmax,pmin, ortabulate. -
- Vector methods
-
-
show,length,c -
Test functions:
is.vector,is.na -
Conversion functions:
as.vector,as.character,as.numeric,as.integer,as.logical -
[,[<-,| -
by,Compare,head,%in%,paste,sort,table,tail,tapply,unique
This release does not support
interaction,lengthb,rank, orsplit. -
Example 5-1 shows simple data preparation and processing. For additional details, see "Support for Hive Data Types."
Example 5-1 Using R to Process Data in Hive Tables
# Connect to Hive ore.connect(type="HIVE") # Attach the current envt. into search path of R ore.attach() # create a Hive table by pushing the numeric columns of the iris data set IRIS_TABLE <- ore.push(iris[1:4]) # Create bins based on Petal Length IRIS_TABLE$PetalBins = ifelse(IRIS_TABLE$Petal.Length < 2.0, "SMALL PETALS", + ifelse(IRIS_TABLE$Petal.Length < 4.0, "MEDIUM PETALS", + ifelse(IRIS_TABLE$Petal.Length < 6.0, + "MEDIUM LARGE PETALS", "LARGE PETALS"))) #PetalBins is now a derived column of the HIVE object > names(IRIS_TABLE) [1] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width" "PetalBins" # Based on the bins, generate summary statistics for each group aggregate(IRIS_TABLE$Petal.Length, by = list(PetalBins = IRIS_TABLE$PetalBins), + FUN = summary) 1 LARGE PETALS 6 6.025000 6.200000 6.354545 6.612500 6.9 0 2 MEDIUM LARGE PETALS 4 4.418750 4.820000 4.888462 5.275000 5.9 0 3 MEDIUM PETALS 3 3.262500 3.550000 3.581818 3.808333 3.9 0 4 SMALL PETALS 1 1.311538 1.407692 1.462000 1.507143 1.9 0 Warning message: ORE object has no unique key - using random order
5.3.3 Support for Hive Data Types
Oracle R Connector for Hadoop can access any Hive table containing columns with string and numeric data types such as tinyint, smallint, bigint, int, float, and double. There is no support for these complex data types:
array binary map struct timestamp union
If you attempt to access a Hive table containing an unsupported data type, then you get an error message. To access the table, you must convert the column to a supported data type.
To convert a column to a supported data type:
-
Open the Hive command interface:
$ hive hive>
-
Identify the column with an unsupported data type:
hive> describe table_name; -
View the data in the column:
hive> select column_name from table_name;
-
Create a table for the converted data, using only supported data types.
-
Copy the data into the new table, using an appropriate conversion tool.
Example 5-2 shows the conversion of an array. Example 5-3 and Example 5-4 show the conversion of timestamp data.
Example 5-2 Converting an Array to String Columns
R> ore.sync(table="t1")
Warning message:
table t1 contains unsupported data types
.
.
.
hive> describe t1;
OK
col1 int
col2 array<string>
hive> select * from t1;
OK
1 ["a","b","c"]
2 ["d","e","f"]
3 ["g","h","i"]
hive> create table t2 (c1 string, c2 string, c2 string);
hive> insert into table t2 select col2[0], col2[1], col2[2] from t1;
.
.
.
R> ore.sync(table="t2")
R> ore.ls()
[1] "t2"
R> t2$c1
[1] "a" "d" "g"
Example 5-3 uses automatic conversion of the timestamp data type into string. The data is stored in a table named t5 with a column named tstmp.
Example 5-3 Converting a Timestamp Column
hive> select * from t5; hive> create table t6 (timestmp string); hive> insert into table t6 SELECT tstmp from t5;
Example 5-4 uses the Hive get_json_object function to extract the two columns of interest from the JSON table into a separate table for use by Oracle R Connector for Hadoop.
Example 5-4 Converting a Timestamp Column in a JSON File
hive> select * from t3;
OK
{"custId":1305981,"movieId":null,"genreId":null,"time":"2010-12-30:23:59:32","recommended":null,"activity":9}
hive> create table t4 (custid int, time string);
hive> insert into table t4 SELECT cast(get_json_object(c1, '$.custId') as int), cast(get_json_object(c1, '$.time') as string) from t3;
5.3.4 Usage Notes for Hive Access
The Hive command language interface (CLI) is used for executing queries and provides support for Linux clients. There is no JDBC or ODBC support.
The ore.create function creates Hive tables only as text files. However, Oracle R Connector for Hadoop can access Hive tables stored as either text files or sequence files.
You can use the ore.exec function to execute Hive commands from the R console. For a demo, run the hive_sequencefile demo.
Oracle R Connector for Hadoop can access tables and views in the default Hive database only. To allow read access to objects in other databases, you must expose them in the default database. For example, you can create views.
Oracle R Connector for Hadoop does not have a concept of ordering in Hive. An R frame persisted in Hive might not have the same ordering after it is pulled out of Hive and into memory. Oracle R Connector for Hadoop is designed primarily to support data cleanup and filtering of huge HDFS data sets, where ordering is not critical. You might see warning messages when working with unordered Hive frames:
Warning messages: 1: ORE object has no unique key - using random order 2: ORE object has no unique key - using random order
To suppress these warnings, set the ore.warn.order option in your R session:
R> options(ore.warn.order = FALSE)
5.3.5 Example: Loading Hive Tables into Oracle R Connector for Hadoop
Table 6-1 provides an example of loading a Hive table into an R data frame for analysis. It uses these Oracle R Connector for Hadoop functions:
hdfs.attach ore.attach ore.connect ore.create ore.hiveOptions ore.sync
Example 5-5 Loading a Hive Table
# Connect to HIVE metastore and sync the HIVE input table into the R session.
ore.connect(type="HIVE")
ore.sync(table="datatab")
ore.attach()
# The "datatab" object is a Hive table with columns named custid, movieid, activity, and rating.
# Perform filtering to remove missing (NA) values from custid and movieid columns
# Project out three columns: custid, movieid and rating
t1 <- datatab[!is.na(datatab$custid) &
!is.na(datatab$movieid) &
datatab$activity==1, c("custid","movieid", "rating")]
# Set HIVE field delimiters to ','. By default, it is Ctrl+a for text files but
# ORCH 2.0 supports only ',' as a file separator.
ore.hiveOptions(delim=',')
# Create another Hive table called "datatab1" after the transformations above.
ore.create (t1, table="datatab1")
# Use the HDFS directory, where the table data for datatab1 is stored, to attach
# it to ORCH framework. By default, this location is "/user/hive/warehouse"
dfs.id <- hdfs.attach("/user/hive/warehouse/datatab1")
# dfs.id can now be used with all hdfs.*, orch.* and hadoop.* APIs of ORCH for further processing and analytics.
5.4 Access to Oracle Database
Oracle R Connector for Hadoop provides a basic level of database access. You can move the contents of a database table to HDFS, and move the results of HDFS analytics back to the database.
You can then perform additional analysis on this smaller set of data using a separate product named Oracle R Enterprise. It enables you to perform statistical analysis on database tables, views, and other data objects using the R language. You have transparent access to database objects, including support for Business Intelligence and in-database analytics.
Access to the data stored in an Oracle database is always restricted to the access rights granted by your DBA.
Oracle R Enterprise is included in the Oracle Advanced Analytics option to Oracle Database Enterprise Edition. It is not one of the Oracle Big Data Connectors.
See Also:
Oracle R Enterprise User's Guide5.4.1 Usage Notes for Oracle Database Access
Oracle R Connector for Hadoop uses Sqoop to move data between HDFS and Oracle Database. Sqoop imposes several limitations on Oracle R Connector for Hadoop:
-
You cannot import Oracle tables with
BINARY_FLOATorBINARY_DOUBLEcolumns. As a work-around, you can create a view that casts these columns toNUMBER. -
All column names must be in upper case.
5.4.2 Scenario for Using Oracle R Connector for Hadoop with Oracle R Enterprise
The following scenario may help you identify opportunities for using Oracle R Connector for Hadoop with Oracle R Enterprise.
Using Oracle R Connector for Hadoop, you can look for files that you have access to on HDFS and execute R calculations on data in one such file. You can also upload data stored in text files on your local file system into HDFS for calculations, schedule an R script for execution on the Hadoop cluster using DBMS_SCHEDULER, and download the results into a local file.
Using Oracle R Enterprise, you can open the R interface and connect to Oracle Database to work on the tables and views that are visible based on your database privileges. You can filter out rows, add derived columns, project new columns, and perform visual and statistical analysis.
Again using Oracle R Connector for Hadoop, you might deploy a MapReduce job on Hadoop for CPU-intensive calculations written in R. The calculation can use data stored in HDFS or, with Oracle R Enterprise, in an Oracle database. You can return the output of the calculation to an Oracle database and to the R console for visualization or additional processing.
5.5 Analytic Functions in Oracle R Connector for Hadoop
Table 5-1 describes the analytic functions. For more information, use R Help.
Table 5-1 Descriptions of the Analytic Functions
| Function | Description |
|---|---|
|
Evaluates a fit generated by |
|
|
Exports a model (W and H factor matrices) to the specified destination for |
|
|
Fits a linear model using tall-and-skinny QR (TSQR) factorization and parallel distribution. The function computes the same statistical parameters as the Oracle R Enterprise |
|
|
Fits a low rank matrix factorization model using either the jellyfish algorithm or the Mahout alternating least squares with weighted regularization (ALS-WR) algorithm. |
|
|
Provides a neural network to model complex, nonlinear relationships between inputs and outputs, or to find patterns in the data. |
|
|
Provides the main entry point to create a nonnegative matrix factorization model using the jellyfish algorithm. This function can work on much larger data sets than the R |
|
|
Plugs in to the R |
|
|
Computes the top n items to be recommended for each user that has predicted ratings based on the input |
|
|
Predicts future results based on the fit calculated by |
|
|
Prints a model returned by the |
|
|
Prints a summary of the fit calculated by |
|
|
Prepares a summary of the fit calculated by |
5.6 ORCH mapred.config Class
The hadoop.exec and hadoop.run functions have an optional argument, config, for configuring the resultant MapReduce job. This argument is an instance of the mapred.config class.
The mapred.config class has these slots:
- hdfs.access
-
Set to
TRUEto allow access to theHDFS.*functions in the mappers, reducers, and combiners, or set toFALSEto restrict access (default). - job.name
-
A descriptive name for the job so that you can monitor its progress more easily.
- map.input
-
A mapper input data-type keyword:
data.frame,list, orvector(default). - map.output
-
A sample R data frame object that defines the output structure of data from the mappers. It is required only if the mappers change the input data format. Then the reducers require a sample data frame to parse the input stream of data generated by the mappers correctly.
If the mappers output exactly the same records as they receive, then you can omit this option.
- map.split
-
The number of rows that your mapper function receives from a mapper.
-
0 sends all rows given by Hadoop to a specific mapper to your mapper function. Use this setting only if you are sure that the chunk of data for each mapper can fit into R memory. If it does not, then the R process will fail with a memory allocation error.
-
1 sends one row only to the mapper at a time (Default). You can improve the performance of a mapper by increasing this value, which decreases the number of invocations of your mapper function. This is particularly important for small functions.
-
n sends a minimum of n rows to the mapper at a time. In this syntax, n is an integer greater than 1. Some algorithms require a minimum number of rows to function.
-
- map.tasks
-
The number of mappers to run. Specify
1to run the mappers sequentially; specify a larger integer to run the mappers in parallel. If you do not specify a value, then the number of mappers is determined by the capacity of the cluster, the workload, and the Hadoop configuration. - map.valkey
-
Set to
TRUEto duplicate the keys as data values for the mapper, orFALSEto use the keys only as keys (default). - min.split.size
-
Controls the lower boundary for splitting HDFS files before sending them to the mappers. This option reflects the value of the Hadoop
mapred.min.split.sizeoption and is set in bytes. - reduce.input
-
A reducer input data type keyword:
data.frameorlist(default). - reduce.output
-
A sample R data frame object that defines the output structure of data from the reducers. This is optional parameter.
The sample data frame is used to generate metadata for the output HDFS objects. It reduces the job execution time by eliminating the sampling and parsing step. It also results in a better output data format, because you can specify column names and other metadata. If you do not specify a sample data frame, then the output HDFS files are sampled and the metadata is generated automatically.
- reduce.split
-
The number of data values given at one time to the reducer. See the values for
map.split. The reducer expects to receive all values at one time, so you must handle partial data sets in the reducer if you setreduce.splitto a value other than 0 (zero). - reduce.tasks
-
The number of reducers to run. Specify
1to run the reducers sequentially; specify a larger integer to run the reducers in parallel. If you do not specify a value, then the number of reducers is determined by the capacity of the cluster, the workload, and the Hadoop configuration. - reduce.valkey
-
Set to
TRUEto duplicate the keys as data values for the reducer, orFALSEto use the keys only as keys (default). - verbose
-
Set to
TRUEto generate diagnostic information, orFALSEotherwise.
5.7 Examples and Demos of Oracle R Connector for Hadoop
The ORCH package includes sample code to help you learn to adapt your R programs to run on a Hadoop cluster using Oracle R Connector for Hadoop. This topic describes these examples and demonstrations.
5.7.1 Using the Demos
Oracle R Connector for Hadoop provides an extensive set of demos. Instructions for running them are included in the following descriptions.
If an error occurs, then exit from R without saving the workspace image and start a new session. You should also delete the temporary files created in both the local file system and the HDFS file system:
# rm -r /tmp/orch* # hadoop fs -rm -r /tmp/orch*
Demo R Programs
- hdfs_cpmv.R
-
Demonstrates copying, moving, and deleting HDFS files and directories.
This program uses the following
ORCHfunctions:hdfs.cd hdfs.cp hdfs.exists hdfs.ls hdfs.mkdir hdfs.mv hdfs.put hdfs.pwd hdfs.rmdir hdfs.root hdfs.setroot
To run this demo, use the following command:
R> demo("hdfs_cpmv", package="ORCH") - hdfs_datatrans.R
-
Demonstrates data transfers between HDFS and the local file system, and between HDFS and an Oracle database.
Note:
This demo requires that Oracle R Enterprise client is installed on Oracle Big Data Appliance, and Oracle R Enterprise server is installed on the Oracle Database host.You must connect to Oracle Database using both Oracle R Connector for Hadoop and Oracle R Enterprise to run this demo to completion. See
orch.connectandore.connecthelp.This program uses the following
ORCHfunctions:hdfs.cd hdfs.describe hdfs.download hdfs.get hdfs.pull hdfs.pwd hdfs.rm hdfs.root hdfs.setroot hdfs.size hdfs.upload orch.connected ore.dropFoot 1 ore.is.connectedFootref 1
To run this demo, use the following commands, replacing the parameters shown here for
ore.connectandorch.connectwith those appropriate for your Oracle database:R> ore.connect("RQUSER", "orcl", "localhost", "welcome1") Loading required package: ROracle Loading required package: DBI R> orch.connect("localhost", "RQUSER", "orcl", "welcome1", secure=F) Connecting ORCH to RDBMS via [sqoop] Host: localhost Port: 1521 SID: orcl User: RQUSER Connected. [1] TRUE R> demo("hdfs_datatrans", package="ORCH")
- hdfs_dir.R
-
Demonstrates the use of functions related to HDFS directories.
This program uses the following
ORCHfunctions:hdfs.cd hdfs.ls hdfs.mkdir hdfs.pwd hdfs.rmdir hdfs.root hdfs.setroot
To run this demo, use the following command:
R> demo("hdfs_dir", package="ORCH") - hdfs_putget.R
-
Demonstrates how to transfer data between an R data frame and HDFS.
This program uses the following
ORCHfunctions:hdfs.attach hdfs.describe hdfs.exists hdfs.get hdfs.put hdfs.pwd hdfs.rm hdfs.root hdfs.sample hdfs.setroot
To run this demo, use the following command:
R> demo("hdfs_putget", package="ORCH") - hive_aggregate.R
-
Moves a selection of the Iris data set into Hive and performs these aggregations for each species: summary, mean, minimum, maximum, standard deviation, median, and interquartile range (IQR).
This program uses the following
ORCHfunctions to set up a Hive table. These functions are used in all of the Hive demo programs and are documented in R Help:ore.attach ore.connect ore.push
To run this demo, use the following command:
R> demo("hive_aggregate", package="ORCH") - hive_analysis.R
-
Demonstrates basic analysis and data processing operations.
To run this demo, use the following command:
R> demo("hive_analysis", package="ORCH") - hive_basic.R
-
Using basic R functions, this demo obtains information about a Hive table, including the number of rows (
nrow), the number of columns (length), the first five rows (head), the class and data type of a column (classandis.*), and the number of characters in each column value (nchar).To run this program, use the following command:
R> demo("hive_basic", package="ORCH") - hive_binning.R
-
Creates bins in Hive based on petal size in the Iris data set.
To run this program, use the following command:
R> demo("hive_binning", package="ORCH") - hive_columnfns.R
-
Uses the Iris data set to show the use of several column functions including
min,max,sd,mean,fivenum,var,IQR,quantile,log,log2,log10,abs, andsqrt.To run this program, use the following command:
R> demo("hive_columnfns", package="ORCH") - hive_nulls.R
-
Demonstrates the differences between handling nulls in R and Hive, using the
AIRQUALITYdata set.This program uses the following
ORCHfunctions, which are documented in R Help:ore.attach ore.connect ore.na.extract ore.push
To run this program, use the following command:
R> demo("hive_nulls", package="ORCH") - hive_pushpull.R
-
Shows processing of the Iris data set split between Hive and the client R desktop.
This program uses the following
ORCHfunctions, which are documented in R Help:ore.attach ore.connect ore.pull ore.push
To run this program, use the following command:
R> demo("hive_pushpull", package="ORCH") - hive_sequencefile.R
-
Shows how to create and use Hive tables stored as sequence files, using the
carsdata set.This program uses the following
ORCHfunctions, which are documented in R Help:ore.attach ore.connect ore.create ore.exec ore.sync ore.drop
To run this program, use the following command:
R> demo("hive_sequencefile", package="ORCH") - mapred_basic.R
-
Provides a simple example of mappers and reducers written in R. It uses the
carsdata set.This program uses the following
ORCHfunctions:hadoop.run hdfs.get hdfs.put hdfs.rm hdfs.setroot orch.keyval
To run this program, use the following command:
R> demo("mapred_basic", package="ORCH") - mapred_modelbuild.R
-
Runs a mapper and a reducer in a parallel model build, saves plots in HDFS in parallel, and displays the graphs. It uses the
irisdata set.This program uses the following
ORCHfunctions:hadoop.run hdfs.download hdfs.exists hdfs.get hdfs.id hdfs.put hdfs.rm hdfs.rmdir hdfs.root hdfs.setroot hdfs.upload orch.export orch.pack orch.unpack
To run this program, use the following command:
R> demo("mapred_modelbuild", package="ORCH") - orch_lm.R
-
Fits a linear model using the
orch.lmalgorithm and compares it with the Rlmalgorithm. It uses theirisdata set.This program uses the following
ORCHfunctions:hdfs.cd hdfs.get hdfs.put hdfs.pwd hdfs.rm hdfs.root hdfs.setroot orch.lmFootref 1 predict.orch.lmFootref 1
To run this program, use the following command:
R> demo("orch_lm", package="ORCH") - orch_lmf_jellyfish.R
-
Fits a low rank matrix factorization model using the jellyfish algorithm.
This program uses the following
ORCHfunctions:hdfs.cd hdfs.get hdfs.mkdir hdfs.pwd hdfs.rmdir hdfs.root hdfs.setroot orch.evaluateFootref 1 orch.export.fitFootref 1 orch.lmfFootref 1
To run this program, use the following command:
R> demo("orch_lmf_jellyfish", package="ORCH") - orch_lmf_mahout_als.R
-
Fits a low rank matrix factorization model using the Mahout ALS-WR algorithm.
This program uses the following
ORCHfunctions:hdfs.cd hdfs.mkdir hdfs.pwd hdfs.rmdir hdfs.root orch.evaluateFootref 1 orch.lmfFootref 1 orch.recommendFootref 1
To run this program, use the following command:
R> demo("orch_lmf_mahout_als", package="ORCH") - orch_neural.R
-
Builds a model using the Oracle R Connector for Hadoop neural network algorithm. It uses the
irisdata set.This program uses the following
ORCHfunctions:hdfs.cd hdfs.put hdfs.pwd hdfs.rm hdfs.root hdfs.sample hdfs.setroot orch.neuralFootref 1
To run this program, use the following command:
R> demo("orch_neural", package="ORCH") - orch_nmf.R
-
Demonstrates the integration of the
orch.nmffunction with the framework of the RNMFpackage to create a nonnegative matrix factorization model.This demo uses the Golub data set provided with the
NMFpackage.This program uses the following
ORCHfunctions:hdfs.cd hdfs.mkdir hdfs.pwd hdfs.rmdir hdfs.root hdfs.setroot orch.nmf.NMFalgoFootref 1
This program requires several R packages that you may not have already installed:
NMF,Biobase,BiocGenerics, andRColorBrewer. Instructions for installing them are provided here.Caution:
When asked whether you want to update the dependent packages, always respond Update/None. The installed R packages are the supported versions. If you update them, then the Oracle Big Data Appliance software checks fail, which causes problems during routine maintenance. Always run thebdacheckswutility after installing new packages. See thebdacheckswutility in the Oracle Big Data Appliance Owner's Guide.If you are using a Hadoop client, then install the packages on the client instead of Oracle Big Data Appliance.
To install NMF:
-
Download
NMFfrom the CRAN archives athttp://cran.r-project.org/src/contrib/Archive/NMF/NMF_0.5.06.tar.gz -
Install
NMFusing a standard installation method:R> install.packages("/full_path/NMF_0.5.06.tar.gz", REPOS=null)
To install Biobase and BiocGenerics from the BioConductor project:
-
Source in the
biocLitepackage:R> source("http://bioconductor.org/biocLite.R") -
Install the
Biobasepackage:R> biocLite("Biobase") -
Install the
BiocGenericspackage:R> biocLite("BiocGenerics")
To install RColorBrewer:
-
Use a standard method of installing a package from CRAN:
R> install.packages("RColorBrewer")
To run the orch_nmf demo:
-
Load the required packages, if they are not already loaded in this session:
R> library("NMF") R> library("Biobase") R> library("BiocGenerics") R> library("RColorBrewer") -
Run the demo:
R> demo("orch_nmf", package="ORCH")
-
- demo-bagged.clust.R
-
Provides an example of bagged clustering using randomly generated values. The mappers perform k-means clustering analysis on a subset of the data and generate centroids for the reducers. The reducers combine the centroids into a hierarchical cluster and store the
hclustobject in HDFS.This program uses the following
ORCHfunctions:hadoop.exec hdfs.put is.hdfs.id orch.export orch.keyval orch.pack orch.unpack
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/demos/demo-bagged.clust.R") Call: c("hclust", "d", "single") Cluster method : single Distance : euclidean Number of objects: 6
- demo-kmeans.R
-
Performs k-means clustering.
This program uses the following
ORCHfunctions:hadoop.exec hdfs.get orch.export orch.keyval orch.pack orch.unpack
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/demos/demo-kmeans.R") centers x y 1 0.3094056 1.32792762 2 1.7374920 -0.06730981 3 -0.6271576 -1.20558920 4 0.2668979 -0.98279426 5 0.4961453 1.11632868 6 -1.7328809 -1.26335598 removing NaNs if any final centroids x y 1 0.666782828 2.36620810 2 1.658441962 0.33922811 3 -0.627157587 -1.20558920 4 -0.007995672 -0.03166983 5 1.334578589 1.41213532 6 -1.732880933 -1.26335598
5.7.2 Using the Examples
The examples show how you use the Oracle R Connector for Hadoop API. You can view them in a text editor after extracting them from the installation archive files. They are located in the ORCH2.1.0/ORCHcore/examples directory. You can run the examples as shown in the following descriptions.
- example-filter1.R
-
Shows how to use key-value pairs. The mapper function selects cars with a distance value greater than 30 from the
carsdata set, and the reducer function calculates the mean distance for each speed.This program uses the following
ORCHfunctions:hadoop.run hdfs.get hdfs.put orch.keyval
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-filter1.R") Running cars filtering and mean example #1: val1 val2 1 10 34.00000 2 13 38.00000 3 14 58.66667 4 15 54.00000 5 16 36.00000 6 17 40.66667 7 18 64.50000 8 19 50.00000 9 20 50.40000 10 22 66.00000 11 23 54.00000 12 24 93.75000 13 25 85.00000
- example-filter2.R
-
Shows how to use values only. The mapper function selects cars with a distance greater than 30 and a speed greater than 14 from the
carsdata set, and the reducer function calculates the mean speed and distance as one value pair.This program uses the following
ORCHfunctions:hadoop.run hdfs.get hdfs.put orch.keyval
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-filter2.R") Running cars filtering and mean example #2: val1 val2 1 59.52 19.72
- example-filter3.R
-
Shows how to load a local file into HDFS. The mapper and reducer functions are the same as example-filter2.R.
This program uses the following
ORCHfunctions:hadoop.run hdfs.download hdfs.upload orch.keyval
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-filter3.R") Running cars filtering and mean example #3: [1] "\t59.52,19.72"
- example-group.apply.R
-
Shows how to build a parallel model and generate a graph. The mapper partitions the data based on the petal lengths in the
irisdata set, and the reducer uses basic R statistics and graphics functions to fit the data into a linear model and plot a graph.This program uses the following
ORCHfunctions:hadoop.run hdfs.download hdfs.exists hdfs.get hdfs.id hdfs.mkdir hdfs.put hdfs.rmdir hdfs.upload orch.pack orch.export orch.unpack
To run this program, ensure that
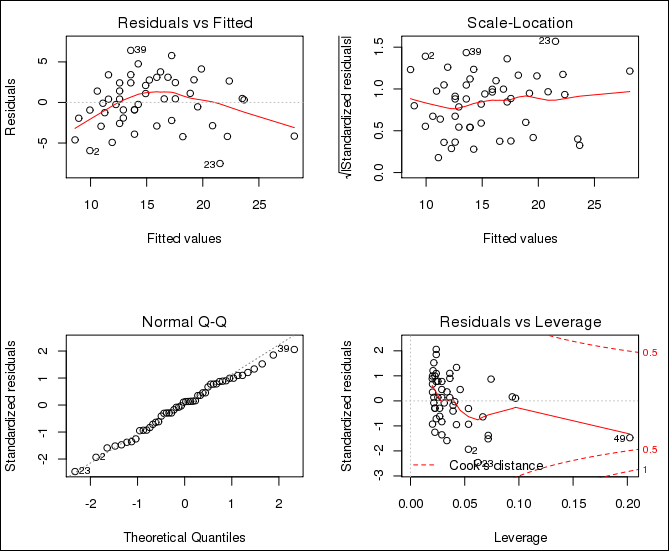
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-group.apply.R") Running groupapply example. [[1]] [[1]]$predict . . . [3] [[3]]$predict 2 210 3 4 5 6 7 8 4.883662 5.130816 4.854156 5.163097 4.109921 4.915944 4.233498 4.886437 9 10 11 12 13 14 15 16 4.789593 4.545214 4.136653 4.574721 4.945451 4.359849 4.013077 4.730579 17 18 19 20 21 22 23 24 4.827423 4.480651 4.792367 4.386581 4.666016 5.007239 5.227660 4.607002 25 26 27 28 29 30 31 32 4.701072 4.236272 4.701072 5.039521 4.604228 4.171709 4.109921 4.015851 33 34 35 36 37 38 39 40 4.574721 4.201216 4.171709 4.139428 4.233498 4.542439 4.233498 5.733065 41 42 43 44 45 46 47 48 4.859705 5.851093 5.074577 5.574432 6.160034 4.115470 5.692460 5.321730 49 50 51 52 53 54 55 56 6.289160 5.386293 5.230435 5.665728 4.891986 5.330054 5.606714 5.198153 57 58 59 60 61 62 63 64 6.315892 6.409962 4.607002 5.915656 4.830198 6.127753 5.074577 5.603939 65 66 67 68 69 70 71 72 5.630671 5.012788 4.951000 5.418574 5.442532 5.848318 6.251329 5.512644 73 74 75 76 77 78 79 80 4.792367 4.574721 6.409962 5.638995 5.136365 4.889212 5.727516 5.886149 81 82 83 84 85 86 87 88 5.915656 4.859705 5.853867 5.980218 5.792079 5.168646 5.386293 5.483137 89 4.827423 [[3]]$pngfile [1] "/user/oracle/pngfiles/3" [1] "/tmp/orch6e295a5a5da9"
This program generates three graphics files in the /tmp directory. Figure 5-1 shows the last one.
Figure 5-1 Example example-group.apply.R Output in fit-3.png
Description of "Figure 5-1 Example example-group.apply.R Output in fit-3.png"
- example-kmeans.R
-
Defines a k-means clustering function and generates random points for a clustering test. The results are printed or graphed.
This program uses the following
ORCHfunctions:hadoop.exec hdfs.get hdfs.put orch.export
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-kmeans.R") Running k-means example. val1 val2 1 1.005255389 1.9247858 2 0.008390976 2.5178661 3 1.999845464 0.4918541 4 0.480725254 0.4872837 5 1.677254045 2.6600670
- example-lm.R
-
Shows how to define multiple mappers and one reducer that merges all results. The program calculates a linear regression using the
irisdata set.This program uses the following
ORCHfunctions:hadoop.run hdfs.get hdfs.put orch.export orch.pack orch.unpack
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-lm.R") Running linear model example. Model rank 3, yy 3.0233000000E+02, nRows 150 Model coefficients -0.2456051 0.2040508 0.5355216
- example-logreg.R
-
Performs a one-dimensional, logistic regression on the
carsdata set.This program uses the following
ORCHfunctions:hadoop.run hdfs.put orch.export
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-logreg.R") Running logistic regression. [1] 1924.1
- example-map.df.R
-
Shows how to run the mapper with an unlimited number of records at one time input as a data frame. The mapper selects cars with a distance greater than 30 from the
carsdata set and calculates the mean distance. The reducer merges the results.This program uses the following
ORCHfunctions:hadoop.run hdfs.get hdfs.put
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-map.df.R")) Running example of data.frame mapper input: val1 val2 1 17.66667 50.16667 2 13.25000 47.25000
- example-map.list.R
-
Shows how to run the mapper with an unlimited number of records at one time input as a list. The mapper selects cars with a distance greater than 30 from the
carsdata set and calculates the mean distance. The reducer merges the results.This program uses the following
ORCHfunctions:hadoop.run hdfs.get hdfs.put
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-map.list.R" Running example of list mapper input: val1 val2 1 15.9 49
- example-model.plot.R
-
Shows how to create models and graphs using HDFS. The mapper provides key-value pairs from the
irisdata set to the reducer. The reducer creates a linear model from data extracted from the data set, plots the results, and saves them in three HDFS files.This program uses the following
ORCHfunctions:hadoop.run hdfs.download hdfs.exists hdfs.get hdfs.id hdfs.mkdir hdfs.put hdfs.rmdir hdfs.upload orch.export orch.pack
To run this program, ensure that
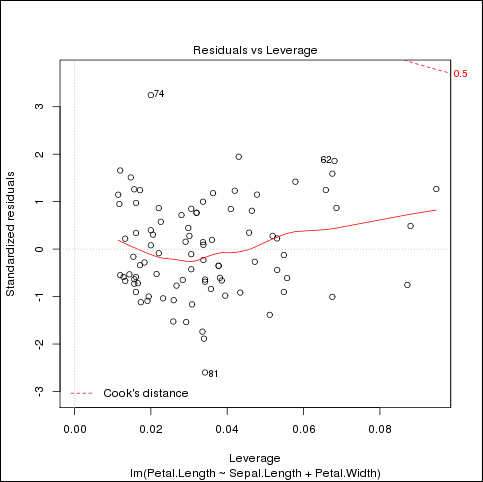
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-model.plot.R" Running model building and graph example. [[1]] [[1]]$predict . . . [[3]]$predict 2 210 3 4 5 6 7 8 5.423893 4.955698 5.938086 5.302979 5.517894 6.302640 4.264954 6.032087 9 10 11 12 13 14 15 16 5.594622 6.080090 5.483347 5.393163 5.719353 4.900061 5.042065 5.462257 17 18 19 20 21 22 23 24 5.448801 6.392824 6.410097 5.032427 5.826811 4.827150 6.358277 5.302979 25 26 27 28 29 30 31 32 5.646442 5.959176 5.230068 5.157157 5.427710 5.924630 6.122271 6.504099 33 34 35 36 37 38 39 40 5.444983 5.251159 5.088064 6.410097 5.406619 5.375890 5.084247 5.792264 41 42 43 44 45 46 47 48 5.698262 5.826811 4.955698 5.753900 5.715536 5.680989 5.320252 5.483347 49 50 5.316435 5.011336 [[3]]$pngfile [1] "/user/oracle/pngfiles/virginica" [1] "/tmp/orch6e29190de160"
The program generates three graphics files. Figure 5-2 shows the last one.
Figure 5-2 Example example-model.plot.R Output in fit-virginica.png
Description of "Figure 5-2 Example example-model.plot.R Output in fit-virginica.png"
- example-model.prep.R
-
Shows how to distribute data across several map tasks. The mapper generates a data frame from a slice of input data from the
irisdata set. The reducer merges the data frames into one output data set.This program uses the following
ORCHfunctions:hadoop.exec hdfs.get hdfs.put orch.export orch.keyvals
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-model.prep.R") v1 v2 v3 v4 1 5.1 4.90 4.055200 0.33647224 2 4.9 4.20 4.055200 0.33647224 3 4.7 4.16 3.669297 0.26236426 . . . 299 6.2 18.36 221.406416 1.68639895 300 5.9 15.30 164.021907 1.62924054
- example-rlm.R
-
Shows how to convert a simple R program into one that can run as a MapReduce job on a Hadoop cluster. In this example, the program calculates and graphs a linear model on the
carsdata set using basic R functions.This program uses the following
ORCHfunctions:hadoop.run orch.keyvals orch.unpack
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-rlm.R" [1] "--- Client lm:" Call: lm(formula = speed ~ dist, data = cars) Residuals: Min 1Q Median 3Q Max -7.5293 -2.1550 0.3615 2.4377 6.4179 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 8.28391 0.87438 9.474 1.44e-12 *** dist 0.16557 0.01749 9.464 1.49e-12 *** --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 3.156 on 48 degrees of freedom Multiple R-squared: 0.6511, Adjusted R-squared: 0.6438 F-statistic: 89.57 on 1 and 48 DF, p-value: 1.49e-12
Enter Ctrl+C to get the R prompt, and then close the graphics window:
R> dev.off()
Figure 5-3 shows the four graphs generated by the program.
- example-split.map.R
-
Shows how to split the data in the mapper. The first job runs the mapper in list mode and splits the list in the mapper. The second job splits a data frame in the mapper. Both jobs use the
carsdata set.This program uses the following
ORCHfunctions:hadoop.run hdfs.get hdfs.put orch.keyval
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-split.map.R") Running example of list splitting in mapper key count splits 1 NA 50 6 Running example of data.frame splitting in mapper key count splits 1 NA 50 8
- example-split.reduce.R
-
Shows how to split the data from the
carsdata set in the reducer.This program uses the following
ORCHfunctions:hadoop.run hdfs.get hdfs.put orch.keyval
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-split.reduce.R") Running example of data.frame reducer input ^C key count 1 1 9 2 1 9 3 1 9 . . . 20 1 9 21 1 5
- example-sum.R
-
Shows how to perform a sum operation in a MapReduce job. The first job sums a vector of numeric values, and the second job sums all columns of a data frame.
This program uses the following
ORCHfunctions:hadoop.run orch.keyval
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-sum.R" val2 1 6 val2 val3 1 770 2149
- example-teragen.matrix.R
-
Shows how to generate large data sets in a matrix for testing programs in Hadoop. The mappers generate samples of random data, and the reducers merge them.
This program uses the following
ORCHfunctions:hadoop.run hdfs.put orch.export orch.keyvals
To run this program, ensure that
rootis set to /tmp, and then source the file. The program runs without printing any output.R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-teragem.matrix.R" Running TeraGen-PCA example: R>
- example-teragen.xy.R
-
Shows how to generate large data sets in a data frame for testing programs in Hadoop. The mappers generate samples of random data, and the reducers merge them.
This program uses the following
ORCHfunctions:hadoop.run hdfs.put orch.export orch.keyvals
To run this program, ensure that
rootis set to /tmp, and then source the file. The program runs without printing any output.R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-teragen2.xy.R") Running TeraGen2 example.R>
- example-teragen2.xy.R
-
Shows how to generate large data sets in a data frame for testing programs in Hadoop. One mapper generates small samples of random data, and the reducers merge them.
This program uses the following
ORCHfunctions:hadoop.run hdfs.put orch.export orch.keyvals
To run this program, ensure that
rootis set to /tmp, and then source the file. The program runs without printing any output.R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-teragem2.xy.R" Running TeraGen2 example. R>
- example-terasort.R
-
Provides an example of a TeraSort job on a set of randomly generated values.
This program uses the following
ORCHfunction:hadoop.run
To run this program, ensure that
rootis set to /tmp, and then source the file:R> hdfs.setroot("/tmp") [1] "/tmp" R> source("/usr/lib64/R/library/ORCHcore/examples/example-terasort.R" Running TeraSort example: val1 1 -0.001467344 2 -0.004471376 3 -0.005928546 4 -0.007001193 5 -0.010587280 6 -0.011636190
5.8 Security Notes for Oracle R Connector for Hadoop
Oracle R Connector for Hadoop can invoke the Sqoop utility to connect to Oracle Database either to extract data or to store results.
Sqoop is a command-line utility for Hadoop that imports and exports data between HDFS or Hive and structured databases. The name Sqoop comes from "SQL to Hadoop." The following explains how Oracle R Connector for Hadoop stores a database user password and sends it to Sqoop.
Oracle R Connector for Hadoop stores a user password only when the user establishes the database connection in a mode that does not require reentering the password each time. The password is stored encrypted in memory. See orch.connect.
Oracle R Connector for Hadoop generates a configuration file for Sqoop and uses it to invoke Sqoop locally. The file contains the user's database password obtained by either prompting the user or from the encrypted in-memory representation. The file has local user access permissions only. The file is created, the permissions are set explicitly, and then the file is open for writing and filled with data.
Sqoop uses the configuration file to generate custom JAR files dynamically for the specific database job and passes the JAR files to the Hadoop client software. The password is stored inside the compiled JAR file; it is not stored in plain text.
The JAR file is transferred to the Hadoop cluster over a network connection. The network connection and the transfer protocol are specific to Hadoop, such as port 5900.
The configuration file is deleted after Sqoop finishes compiling its JAR files and starts its own Hadoop jobs.
Footnote Legend
Footnote 1: Use the Rhelp function for usage information