6 Using Oracle R Enterprise Embedded R Execution
Embedded R execution is a significant feature of Oracle R Enterprise. This chapter discusses embedded R execution in the following topics:
About Oracle R Enterprise Embedded R Execution
In Oracle R Enterprise, embedded R execution is the ability to store R scripts in Oracle Database and to invoke such scripts, which then execute in one or more R engines that run in the database and that are dynamically started and managed by the database. Oracle R Enterprise provides both an R interface and a SQL interface for embedded R execution. From the same R script you can get structured data, an XML representation of R objects and images, and even PNG images through a BLOB column in a database table.
This section has the following topics:
Benefits of Embedded R Execution
Embedded R execution has the following benefits:
-
Eliminates moving data from the Oracle Database server to your local R session.
As well as being more secure, the transfer of database data between Oracle Database and an internal R engine is much faster than to a separate client R engine.
-
Uses the database server to start, manage, and control the execution of R scripts in database-side R engines.
-
Leverages the memory and processing power of the database server machine for R engine execution, which provides better scalability and performance.
-
Enables data-parallel and task-parallel execution of user-defined R functions that correspond to special cases of Hadoop Map-Reduce jobs.
-
Provides parallel simulations capability.
-
Allows the use of open source CRAN packages at the database server machine.
-
Provides the ability to develop and operationalize comprehensive scripts for analytical applications in a single step, without leaving the R environment.
You can directly integrate R scripts used in exploratory analysis into application tasks. You can also immediately invoke R scripts in production to drastically reduce time to market by eliminating porting and enabling instantaneous updates of changes to application code.
-
Executing R scripts from SQL enables integration of R script results with Oracle Business Intelligence Enterprise Edition (OBIEE), Oracle BI Publisher, and other SQL-enabled tools for structured data, R objects, and images.
APIs for Embedded R Execution
Oracle R Enterprise provides R and SQL application programming interfaces for embedded R execution. Table 6-1 provides a summary of the embedded R execution functions and the R script repository functions available. The function f refers to the user-defined R code, or script, that is provided as either an R function object or a named R function in the database R script repository.
Table 6-1 R and SQL APIs for Embedded R Execution
| R API | SQL API | Description |
|---|---|---|
|
|
|
Executes f with no automatic transfer of data. |
|
|
|
Executes f by passing all rows of the provided input |
|
|
rqGroupEval This function must be explicitly defined by the user. |
Executes f by partitioning data according to the values of a grouping column. Provides each data partition as a |
|
|
|
Executes f by passing a specified number of rows (a chunk) of the provided input |
|
|
No equivalent. |
Executes f with no automatic transfer of data, but provides the index of the invocation, 1 through n, where n is the number of functions to invoke. Supports parallel execution of each f invocation in the pool of database server-side R engines. |
|
|
|
Loads the provided R function into the R script repository with the provided name. |
|
|
|
Removes the named R function from the R script repository. |
Security Considerations for Scripts
Because both R scripts and SQL scripts allow access to the database server, the creation of scripts must be controlled. The RQADMIN role is a collection of Oracle Database privileges that a user must have to create scripts and store them in the Oracle Database R script repository or drop scripts from the repository.
The installation of Oracle R Enterprise creates the RQADMIN role. The role must be explicitly granted to a user. To grant RQADMIN to a user, start SQL*Plus as sysdba and enter a GRANT statement such as the following, which grants the role to the user RQUSER:
GRANT RQADMIN to RQUSER
Note:
You should grant RQADMIN only to those users who need it.Support for Parallel Execution
Some of the Oracle R Enterprise embedded R execution functions support the use of parallel execution in the database. The ore.groupApply and ore.rowApply functions support data-parallel execution and the ore.indexApply function supports task-parallel execution. This parallel execution capability enables a script to take advantage of high-performance computing hardware such as an Oracle Exadata Database Machine.
The parallel argument of these functions specifies the degree of parallelism to use in the embedded R execution. The value of the argument can be one of the following:
-
A positive integer greater than or equal to
2for a specific degree of parallelism -
FALSEor1for no parallelism -
TRUEfor the default parallelism of thedataargument -
NULLfor the database default for the operation
The default value of the argument is the value of the global option ore.parallel or FALSE if ore.parallel is not set.
A user-defined R function invoked using ore.doEval or ore.tableApply is not executed in parallel. The function executes in a single R engine.
In data-parallel execution for the ore.groupApply function, one or more R engines perform the same R function, or task, on different partitions of data. This functionality enables the building of large numbers of models, for example building tens or hundreds of thousands of predictive models, one model per customer.
In data-parallel execution for the ore.rowApply function, one or more R engines performs the same R function on disjoint chunks of data. This functionality enables scalable model scoring and predictions on large data sets.
In task-parallel execution for the ore.indexApply function, one or more R engines perform the same or different calculations, or task. A number, associated with the index of the execution, is provided to the function. This functionality is valuable in a variety of operations, such as in performing simulations.
Oracle Database handles the management and control of potentially multiple R engines at the database server, automatically partitioning and passing data to R engines executing in parallel. It ensures that all of the R function executions for all of the partitions complete; if not, the Oracle R Enterprise function returns an error. The result from the execution of each user-defined embedded R function is gathered in an ore.list. This list remains in the database until the user requires the result.
Embedded R execution also allows for data-parallel execution of user-defined R functions that may use functions from an open source R package from the Comprehensive R Archive Network (CRAN) or other third-party R package. However, third-party packages do not leverage in-database parallelism and are subject to the parallelism constraints of R. Third-party packages can benefit from the data-parallel and task-parallel execution supported in embedded R execution.
See Also:
"Oracle R Enterprise Global Options"Installing a Third-Party Package for Use in Embedded R Execution
Embedded R execution allows the use of CRAN or other third-party packages in user-defined R functions executed on the Oracle Database server. To use a third-party package in embedded R execution, the package must be installed on the database server. If you are going to use the package from the R interface for embedded R execution, then the package must also be installed on the client, as well.
Embedded R execution enables user-defined R functions to use CRAN packages; however, open source R packages do not leverage in-database parallelism and are subject to the parallelism constraints of R. CRAN packages can benefit through the data-parallel and task-parallel execution supported by embedded R execution.
Embedded R execution leverages what is likely a larger amount of memory and number of processors on the database server, such as an Oracle Exadata Database Machine, than is available on a typical R client machine. Embedded R execution provides for a more efficient transfer of data between the database and the R engine because they are on the same machine.
You can install a third-party package in an R session or from the command line. For information on installing a package in an R session, see "Using a Third-Party Package on the Client".
To install a package on the server so that it can be used by any R user and for use in embedded R execution, an Oracle Database Administrator (DBA) typically executes commands from the command line or in an administrative tool.
A DBA would typically do the following:
-
Download the package source from CRAN using
wget. If the package depends on any packages that are not in the R distribution in use, then download the sources for those packages, also. -
Do one of the following:
-
For a single Oracle Database instance, use the
ORE CMD INSTALLcommand to install the package or packages in the same location as the Oracle R Enterprise packages, which is$ORACLE_HOME/R/library. -
For installing a package on multiple database servers, such as those in an Oracle Real Application Clusters (Oracle RAC) or a multinode Oracle Exadata Database Machine environment, use the Exadata Distributed Command Line Interface (DCLI) utility. To install a package on a single node, use the
ORE CMD INSTALLcommand.
-
Example 6-1 demonstrates getting the source for the arules package from CRAN and installing it with ORE CMD INSTALL from a Linux command line.
Example 6-1 Installing a Package for a Single Database
wget http://cran.r-project.org/src/contrib/arules_1.1-1.tar.gz ORE CMD INSTALL arules_1.1-1.tar.gz
Example 6-2 shows the DLCI command for installing the arules package.
R Interface for Embedded R Execution
Oracle R Enterprise provides functions that invoke R scripts that run in one or more R engines that are embedded in the Oracle database. Other functions create R scripts that are stored in a database script repository or drop a script from the repository. This section describes those functions. It contains the following topics:
Arguments for Functions that Run Scripts
The Oracle R Enterprise embedded R execution functions ore.doEval, ore.tableApply, ore.groupApply, ore.rowApply, and ore.indexApply have arguments that are common to some or all of the functions. Some of the functions also have an argument that is unique to the function.
This section describes the arguments in the following topics:
See Also:
-
For function signatures and more details about function arguments, see the online help displayed by invoking
help(ore.doEval) -
For examples of the use of the arguments, see "Using the ore.doEval Function" and the other topics on using the embedded R execution functions.
Input Function to Execute
The embedded R execution functions all require a function to apply during the execution of the script. You specify the input function with one of the following mutually exclusive arguments:
-
FUN -
FUN.NAME
The FUN argument takes a function object as a directly specified function or as one assigned to an R variable. Only a user with the RQADMIN role can use the FUN argument.when invoking an embedded R function.
The FUN.NAME argument specifies a script that is stored in the R script repository. A stored script contains the function to apply when the script runs. Any Oracle R Enterprise user can use the FUN.NAME argument when invoking an embedded R function.
The advanced Oracle R Enterprise analytics functions in the OREmodels package, ore.glm, ore.lm, and ore.neural, use the embedded R execution framework internally and cannot be used in embedded R execution functions.
Optional Arguments
All of the embedded R execution functions also take optional arguments, which can be named or not. Oracle R Enterprise passes user-defined optional arguments to the input function. You can pass any number of optional arguments to the input function, including complex R objects such as models.
Arguments that start with ore. are special control arguments. Oracle R Enterprise does not pass them to the input function, but instead uses them to control what happens before or after the execution of that function. The following control arguments are supported:
-
ore.connectcontrols whether to automatically connect to Oracle R Enterprise inside the embedded R execution function. This is equivalent to doing anore.connectcall with the same credentials as the client session. The default value isFALSE. -
ore.dropcontrols the input data. IfTRUE, a one columndata.frameis converted to a vector. The default value isTRUE. -
ore.na.omitcontrols the handling of missing values in the input data. IfTRUE, rows or vector elements, depending on theore.dropsetting, that contain missing values are removed from the input data. If all of the rows in a chunk contain missing values, then the input data for that chunk will be an emptydata.frameorvector. The default value isFALSE. -
ore.graphicscontrols whether to start a graphical driver and look for images. The default value isTRUE. -
ore.png.*specifies additional arguments for thepnggraphics driver ifore.graphicsisTRUE. The naming convention for these arguments is to add anore.png.prefix to the arguments of thepngfunction. For example, ifore.png.heightis supplied, argumentheightwill be passed to thepngfunction. If not set, the standard default values for thepngfunction are used.
Structure of Return Value
Another argument that applies to all of the embedded R execution functions is FUN.VALUE. If the FUN.VALUE argument is NULL, then the ore.doEval and ore.tableApply function can return a serialized R object as an ore.object class object, and the ore.groupApply, ore.indexApply, and ore.rowApply functions return an ore.list object. However, if you specify a data.frame or an ore.frame with the FUN.VALUE argument, then the function returns an ore.frame that has the structure of the specified data.frame or ore.frame object.
Input Data
The ore.doEval,and ore.indexApply functions do not automatically receive any data from the database. They simply execute the function specified by the FUN or FUN.NAME argument. Any data needed by the input function is either generated within that function or explicitly retrieved from a data source such as Oracle Database, other databases, or flat files. The input function can load data from a file or a table using the ore.pull function or other transparency layer function.
The ore.tableApply, ore.groupApply, and ore.rowApply functions require a database table as input data. The table is represented by an ore.frame. You supply that data with an ore.frame object that you specify with the X argument, which is the first argument to the embedded R execution function. The embedded R execution function passes the ore.frame object to the user-defined input function as the first argument to that function.
Note:
The data represented by theore.frame object passed to the user-defined R function is copied from Oracle Database to the database server R engine. The R memory limitations apply. If your database server machine has 32 GB RAM and your data table is 64 GB, then Oracle R Enterprise cannot load the data into the R engine memory.The embedded R execution function passes the ore.frame object input data provided as the first argument to a user-defined R function invoked using ore.tableApply is physically being moved from Oracle Database to the database server R engine. Note that the R memory limitations still apply. If your database server machine has 32 GB RAM and your data table is 64 GB, Oracle R Enterprise cannot load the data into the R engine memory.
Parallel Execution
The ore.groupApply, ore.indexApply, and ore.rowApply functions take the parallel argument. That argument specifies the degree of parallelism to use in the embedded R execution of the input function. See "Support for Parallel Execution".
Unique Arguments
The ore.groupApply, ore.indexApply, and ore.rowApply functions each take a argument unique to the function.
The ore.groupApply function takes the INDEX argument, which specifies the name of a column by which the rows of the input data are partitioned for processing by the input function.
The ore.indexApply function takes the times argument, which specifies the number of times to execute the input function.
The ore.rowApply function tales the rows argument, which specifies the number of rows to pass to each invocation of the input function.
Automatic Database Connection in Embedded R Scripts
An embedded R script can automatically connect to an Oracle database.
If automatic connections are enabled, the following functionality occurs:
-
Embedded R scripts are automatically connected to the database.
-
The automatic connection has the same credentials as the session that invokes the embedded R SQL functions.
-
The script runs in an autonomous transaction.
-
ROracle queries work with the automatic connection.
-
Oracle R Enterprise transparency is enabled in the embedded script.
-
User and site-wide R profile loading is disabled in embedded R.
Profile loading was supported in earlier Oracle R Enterprise releases. An automatic connection provides a more secure connection.
Automatic connections are disabled by default. You can specify whether automatic connections are enabled or disabled by using the ore.connect control argument. Control arguments are documented in R online Help for ore.doEval.
To enable automatic connections, ROracle was extended by adding a new driver ExtDriver with the constructor Extproc that is initialized by passing an external pointer wrapping the extproc context. Similarly to OraDriver, ExtDriver is a singleton. Both drivers can exist simultaneously in a session because they are represented by two distinct singletons. This setup allows working with extproc and explicit OraDriver connections in the same R script as shown by the following example.
ore.doEval(function() {
ore.disconnect()
con1 <- dbConnect(Extproc())
res1 <- dbGetQuery(con1, "select * from grade order by name")
con2 <- dbConnect(Oracle(), "scott", "tiger")
res2 <- dbGetQuery(con2, "select * from emp order by empno")
dbDisconnect(con1)
dbDisconnect(con2)
cbind(head(res1)[,1:3], head(res2)[,1:3])
} }, ore.connect = TRUE)
Using the ore.doEval Function
The ore.doEval function executes the specified input function using data that is generated by the input function. It returns an ore.frame object or a serialized R object as an ore.object object.
Example 6-3 creates the function object RandomRedDots that gets a function that has an argument and that returns a data.frame object that has two columns and that plots 100 random normal values. The example then invokes ore.doEval function and passes it the RandomRedDots object. The image is displayed at the client, but it is generated by the database server R engine that executed the RandomRedDots function.
Example 6-3 Using the ore.doEval Function
RandomRedDots <- function(divisor=100){
id<- 1:10
plot(1:100, rnorm(100), pch = 21, bg = "red", cex = 2 )
data.frame(id=id, val=id / divisor)
}
ore.doEval(RandomRedDots)
Listing for Example 6-3
R> RandomRedDots <- function(divisor=100){
+ id<- 1:10
+ plot(1:100, rnorm(100), pch = 21, bg = "red", cex = 2 )
+ data.frame(id=id, val=id / divisor)
+ }
R> ore.doEval(RandomRedDots)
id val
1 1 0.01
2 2 0.02
3 3 0.03
4 4 0.04
5 5 0.05
6 6 0.06
7 7 0.07
8 8 0.08
9 9 0.09
10 10 0.10
You can provide arguments to the input function as optional arguments to the doEval function. Example 6-4 invokes the doEval function with an optional argument that overrides the divisor argument of the RandomRedDots function.
Example 6-4 Using the ore.doEval Function with an Optional Argument
ore.doEval(RandomRedDots, divisor=50)
Listing for Example 6-4
R> ore.doEval(RandomRedDots, divisor=50) id val 1 1 0.02 2 2 0.04 3 3 0.06 4 4 0.08 5 5 0.10 6 6 0.12 7 7 0.14 8 8 0.16 9 9 0.18 10 10 0.20 # The graph displayed by the plot function is not shown.
If the input function is stored in the R script repository, then you can invoke the ore.doEval function with the FUN.NAME argument. Example 6-5 invokes the ore.doEval function and specifies myRandomRedDots, which is the RandomRedDots function that was added to the R script repository by that name. The result is assigned to the variable res.
The return value of the RandomRedDots function is a data.frame but in Example 6-5 the ore.doEval function returns an ore.object object. To get back the data.frame object, the example invokes ore.pull to pull the result to the client R engine.
Example 6-5 Using the ore.doEval Function with the FUN.NAME Argument
res <- ore.doEval(FUN.NAME="myRandomRedDots",divisor=50) class(res) res.local <- ore.pull(res) class(res.local)
Listing for Example 6-5
R> res <- ore.doEval(FUN.NAME = "myRandomRedDots", divisor = 50) R> class(res) [1] "ore.object" attr(,"package") [1] "OREembed" R> res.local <- ore.pull(res) R> class(res.local) [1] "data.frame"
To have the doEval function return an ore.frame object instead of an ore.object, specify the argument FUN.VALUE to describes the structure of the result, as shown in Example 6-6.
Example 6-6 Using the ore.doEval Function with the FUN.VALUE Argument
res.of <- ore.doEval(FUN.NAME="myRandomRedDots", divisor=50,
FUN.VALUE= data.frame(id=1, val=1))
class(res.of)
Listing for Example 6-6
R> res.of <- ore.doEval(FUN.NAME="myRandomRedDots", divisor=50, + FUN.VALUE= data.frame(id=1, val=1)) R> class(res.of) [1] "ore.frame" attr(,"package") [1] "OREbase"
To establish a connection to Oracle Database within the input function, set the special optional argument ore.connect TRUE. This uses the credentials of the user who invoked the function ore.doEval to establish a connection and also automatically load the Oracle R Enterprise package. This capability can be useful to explicitly use the Oracle R Enterprise transparency layer or to save and load objects from an Oracle R Enterprise datastore.
Example 6-7 creates the RandomRedDots function object as in Example 6-3 but this time the function has an argument that takes the name of a datastore. The example creates the myVar variable and saves it in the datastore named datastore_1. The example then invokes the doEval function and passes it the name of the datastore and specifies the ore.connect argument.
Example 6-7 Using the doEval Function with the ore.connect Argument
RandomRedDots <- function(divisor=100, datastore.name="myDatastore"){
id <- 1:10
plot(1:100, rnorm(100), pch = 21, bg = "red", cex = 2 )
ore.load(datastore.name) # contains numeric variable myVar
data.frame(id=id, val=id / divisor, num=myVar)
}
myVar <- 5
ore.save(myVar, name = "datastore_1")
ore.doEval(RandomRedDots, datastore.name="datastore_1", ore.connect=TRUE)
Listing for Example 6-7
R> RandomRedDots <- function(divisor=100, datastore.name="myDatastore"){
+ id <- 1:10
+ plot(1:100, rnorm(100), pch = 21, bg = "red", cex = 2 )
+ ore.load(datastore.name) # contains numeric variable myVar
+ data.frame(id=id, val=id / divisor, num=myVar)
+ }
R> ore.doEval(RandomRedDots, datastore.name="datastore_1", ore.connect=TRUE)
id val num
1 1 0.01 5
2 2 0.02 5
3 3 0.03 5
4 4 0.04 5
5 5 0.05 5
6 6 0.06 5
7 7 0.07 5
8 8 0.08 5
9 9 0.09 5
10 10 0.10 5
# The graph displayed by the plot function is not shown.
Using the ore.tableApply Function
The ore.tableApply function invokes an R script with an ore.frame as the input data. The ore.tableApply function passes the ore.frame to the user-defined input function as the first argument to that function. The ore.tableApply function returns an ore.frame object or a serialized R object as an ore.object object.
See Also:
Example 6-8 uses the ore.tableApply function to build a Naive Bayes model on the iris data set. The naiveBayes function is in the e1071 package, which must be installed on both the client and database server machine R engines. As the first argument to the ore.tableApply function, the ore.push(iris) invocation creates a temporary database table and an ore.frame that is a proxy for the table. The second argument is the input function, which has as an argument dat. The ore.tableApply function passes the ore.frame to the input function as the dat argument. The input function creates a model, which the ore.tableApply function returns as an ore.object object.
Example 6-8 Using the ore.tableApply Function
library(e1071)
mod <- ore.tableApply(
ore.push(iris),
function(dat) {
library(e1071)
dat$Species <- as.factor(dat$Species)
naiveBayes(Species ~ ., dat)
})
class(mod)
mod
Listing for Example 6-8
R> mod <- ore.tableApply(
+ ore.push(iris),
+ function(dat) {
+ library(e1071)
+ dat$Species <- as.factor(dat$Species)
+ naiveBayes(Species ~ ., dat)
+ })
R> class(mod)
[1] "ore.object"
attr(,"package")
[1] "OREembed"
R> mod
Naive Bayes Classifier for Discrete Predictors
Call:
naiveBayes.default(x = X, y = Y, laplace = laplace)
A-priori probabilities:
Y
setosa versicolor virginica
0.3333333 0.3333333 0.3333333
Conditional probabilities:
Sepal.Length
Y [,1] [,2]
setosa 5.006 0.3524897
versicolor 5.936 0.5161711
virginica 6.588 0.6358796
Sepal.Width
Y [,1] [,2]
setosa 3.428 0.3790644
versicolor 2.770 0.3137983
virginica 2.974 0.3224966
Petal.Length
Y [,1] [,2]
setosa 1.462 0.1736640
versicolor 4.260 0.4699110
virginica 5.552 0.5518947
Petal.Width
Y [,1] [,2]
setosa 0.246 0.1053856
versicolor 1.326 0.1977527
virginica 2.026 0.2746501
Using the ore.groupApply Function
The ore.groupApply function invokes an R script with an ore.frame as the input data. The ore.groupApply function passes the ore.frame to the user-defined input function as the first argument to that function. The INDEX argument to the ore.groupApply function specifies the name of a column of the ore.frame by which Oracle Database partitions the rows for processing by the user-defined R function. The ore.groupApply function can use data-parallel execution, in which one or more R engines perform the same R function, or task, on different partitions of data.
The ore.groupApply function returns an ore.list object or an ore.frame object.
Examples of the use of the ore.groupApply function are in the following topics:
See Also:
Partitioning on a Single Column
Example 6-9 uses the C50 package to build a C5.0 decision tree model on the churn data set from C50, with the goal of building one churn model on the data for each state. The example does the following:
-
Loads the
C50package and then thechurndata set. -
Uses the
ore.createfunction to create a database table and the proxyore.frameobject fromchurnTrain, adata.frameobject. -
Specifies
CHURN_TRAIN, anore.frameobject, as the first argument to theore.groupApplyfunction and specifies thestatecolumn as theINDEXargument. Theore.groupApplyfunction partitions the data on thestatecolumn and invokes the user-defined function on each partition. -
Specifies the user-defined function. The first argument of the user-defined function receives one partition of the data, which in this case is all of the data associated with a single state.
-
The user-defined function does the following:
-
Loads the C50 package so that it is available to the function when it executes in an R engine in the database.
-
Deletes the
statecolumn from thedata.frameso that the column is not included in the model. -
Convert the columns to factors because, although the
ore.framedefined factors, when they are loaded to the user-defined function, factors appear as character vectors. -
Builds a model for a state and returns it.
-
-
The
ore.groupApplyfunction returns a list that contains the results from the execution of the user-defined function on each partition of the data. In this case, it is one C5.0 model per state. -
The example creates the variable
modList, which gets theore.listobject returned by theore.groupApplyfunction. Theore.listobject contains the results from the execution of the user-defined function on each partition of the data. In this case, it is one C5.0 model per state, each model stored asore.objectobject. -
Uses the
ore.pullfunction to retrieve the model from the database as themod.MAvariable and then invokes thesummaryfunction on it. The class ofmod.MAisC5.0.
Example 6-9 Using the ore.groupApply Function
library(C50)
data("churn")
ore.create(churnTrain, "CHURN_TRAIN")
modList <- ore.groupApply(
CHURN_TRAIN,
INDEX=CHURN_TRAIN$state,
function(dat) {
library(C50)
dat$state <- NULL
dat$churn <- as.factor(dat$churn)
dat$area_code <- as.factor(dat$area_code)
dat$international_plan <- as.factor(dat$international_plan)
dat$voice_mail_plan <- as.factor(dat$voice_mail_plan)
C5.0(churn ~ ., data = dat, rules = TRUE)
});
mod.MA <- ore.pull(modList$MA)
summary(mod.MA)
Listing for Example 6-9
R> library(C50)
R> data(churn)
R>
R> ore.create(churnTrain, "CHURN_TRAIN")
R>
R> modList <- ore.groupApply(
+ CHURN_TRAIN,
+ INDEX=CHURN_TRAIN$state,
+ function(dat) {
+ library(C50)
+ dat$state <- NULL
+ dat$churn <- as.factor(dat$churn)
+ dat$area_code <- as.factor(dat$area_code)
+ dat$international_plan <- as.factor(dat$international_plan)
+ dat$voice_mail_plan <- as.factor(dat$voice_mail_plan)
+ C5.0(churn ~ ., data = dat, rules = TRUE)
+ });
R> mod.MA <- ore.pull(modList$MA)
R> summary(mod.MA)
Call:
C5.0.formula(formula = churn ~ ., data = dat, rules = TRUE)
C5.0 [Release 2.07 GPL Edition] Thu Feb 13 15:09:10 2014
-------------------------------
Class specified by attribute `outcome'
Read 65 cases (19 attributes) from undefined.data
Rules:
Rule 1: (52/1, lift 1.2)
international_plan = no
total_day_charge <= 43.04
-> class no [0.963]
Rule 2: (5, lift 5.1)
total_day_charge > 43.04
-> class yes [0.857]
Rule 3: (6/1, lift 4.4)
area_code in {area_code_408, area_code_415}
international_plan = yes
-> class yes [0.750]
Default class: no
Evaluation on training data (65 cases):
Rules
----------------
No Errors
3 2( 3.1%) <<
(a) (b) <-classified as
---- ----
53 1 (a): class no
1 10 (b): class yes
Attribute usage:
89.23% international_plan
87.69% total_day_charge
9.23% area_code
Time: 0.0 secs
Partitioning on Multiple Columns
The ore.groupApply function takes only a single column for the INDEX argument; however, you can create a new column that is the concatenation of the columns you want to use and provide this new column to the INDEX argument.
Example 6-10 uses data from the CHURN_TRAIN data set to build an rpart model that produces rules on the partitions of data specified, which are the voice_mail_plan and international_plan columns. The example uses the R table function to show the number of rows to expect in each partition. It then adds a new column that pastes together the two columns of interest to create a new column named vmp_ip.g1
The example next invokes the ore.scriptDrop to ensure that no script by the specified name exists in the R script repository. It then uses the ore.scriptCreate function to define a script named my.rpartFunction and to store it in the repository. The stored script defines a function that takes a data source and a prefix to use for naming Oracle R Enterprise datastore objects. Each invocation of the function my.rpartFunction receives data from one of the partitions identified in vmp_ip. Because the source partition columns are constants, the function sets them to NULL. It converts the character vectors to factors, builds a to predict churn, and saves in it an appropriately named datastore. The function creates a list to return the specific partition column values, the distribution of churn values, and the model itself.
The example then loads the rpart library, sets the datastore prefix, and invokes ore.groupApply using the derived column vmp_ip as the input to argument INDEX and my.rpartFunction as the input to argument FUN.NAME to invoke the user-defined function stored in the R script repository. The ore.groupApply function uses an optional argument to pass the datastorePrefix variable to the user-defined function. It uses the optional argument ore.connect to connect to the database when executing the user-defined function. The ore.groupApply function returns an ore.list object as the variable res.
The example displays the first entry in the list returned. It then invokes the ore.load function to load the model for the case where the customer has both the voice mail plan and the international plan.
Example 6-10 Using ore.groupApply for Partitioning Data on Multiple Columns
library(C50)
data(churn)
ore.drop("CHURN_TRAIN")
ore.create(churnTrain, "CHURN_TRAIN")
table(CHURN_TRAIN$international_plan, CHURN_TRAIN$voice_mail_plan)
CT <- CHURN_TRAIN
CT$vmp_ip <- paste(CT$voice_mail_plan,CT$international_plan,sep="-")
options(width = 80)
head(CT, 3)
ore.scriptDrop("my.rpartFunction")
ore.scriptCreate("my.rpartFunction",
function(dat,datastorePrefix) {
library(rpart)
vmp <- dat[1,"voice_mail_plan"]
ip <- dat[1,"international_plan"]
datastoreName <- paste(datastorePrefix,vmp,ip,sep="_")
dat$voice_mail_plan <- NULL
dat$international_plan <- NULL
dat$state <- as.factor(dat$state)
dat$churn <- as.factor(dat$churn)
dat$area_code <- as.factor(dat$area_code)
mod <- rpart(churn ~ ., data = dat)
ore.save(mod, name=datastoreName, overwrite=TRUE)
list(voice_mail_plan=vmp,
international_plan=ip,
churn.table=table(dat$churn),
rpart.model = mod)
})
library(rpart)
datastorePrefix="my.rpartModel"
res <- ore.groupApply( CT, INDEX=CT$vmp_ip,
FUN.NAME="my.rpartFunction",
datastorePrefix=datastorePrefix,
ore.connect=TRUE)
res[[1]]
ore.load(name=paste(datastorePrefix,"yes","yes",sep="_"))
mod
Listing for Example 6-10
R> library(C50)
R> data(churn)
R> ore.drop("CHURN_TRAIN")
R> ore.create(churnTrain, "CHURN_TRAIN")
R>
R> table(CHURN_TRAIN$international_plan, CHURN_TRAIN$voice_mail_plan)
no yes
no 2180 830
yes 231 92
R> CT <- CHURN_TRAIN
R> CT$vmp_ip <- paste(CT$voice_mail_plan,CT$international_plan,sep="-")
R> options(width = 80)
R> head(CT, 3)
state account_length area_code international_plan voice_mail_plan
1 KS 128 area_code_415 no yes
2 OH 107 area_code_415 no yes
3 NJ 137 area_code_415 no no
number_vmail_messages total_day_minutes total_day_calls total_day_charge
1 25 265.1 110 45.07
2 26 161.6 123 27.47
3 0 243.4 114 41.38
total_eve_minutes total_eve_calls total_eve_charge total_night_minutes
1 197.4 99 16.78 244.7
2 195.5 103 16.62 254.4
3 121.2 110 10.30 162.6
total_night_calls total_night_charge total_intl_minutes total_intl_calls
1 91 11.01 10.0 3
2 103 11.45 13.7 3
3 104 7.32 12.2 5
total_intl_charge number_customer_service_calls churn vmp_ip
1 2.70 1 no yes-no
2 3.70 1 no yes-no
3 3.29 0 no no-no
R>
R> ore.scriptDrop("my.rpartFunction")
R> ore.scriptCreate("my.rpartFunction",
+ function(dat,datastorePrefix) {
+ library(rpart)
+ vmp <- dat[1,"voice_mail_plan"]
+ ip <- dat[1,"international_plan"]
+ datastoreName <- paste(datastorePrefix,vmp,ip,sep="_")
+ dat$voice_mail_plan <- NULL
+ dat$international_plan <- NULL
+ dat$state <- as.factor(dat$state)
+ dat$churn <- as.factor(dat$churn)
+ dat$area_code <- as.factor(dat$area_code)
+ mod <- rpart(churn ~ ., data = dat)
+ ore.save(mod, name=datastoreName, overwrite=TRUE)
+ list(voice_mail_plan=vmp,
+ international_plan=ip,
+ churn.table=table(dat$churn),
+ rpart.model = mod)
+ })
R>
R> library(rpart)
R> datastorePrefix="my.rpartModel"
R>
R> res <- ore.groupApply( CT, INDEX=CT$vmp_ip,
+ FUN.NAME="my.rpartFunction",
+ datastorePrefix=datastorePrefix,
+ ore.connect=TRUE)
R> res[[1]]
$voice_mail_plan
[1] "no"
$international_plan
[1] "no"
$churn.table
no yes
1878 302
$rpart.model
n= 2180
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 2180 302 no (0.86146789 0.13853211)
2) total_day_minutes< 263.55 2040 192 no (0.90588235 0.09411765)
4) number_customer_service_calls< 3.5 1876 108 no (0.94243070 0.05756930)
8) total_day_minutes< 223.25 1599 44 no (0.97248280 0.02751720) *
9) total_day_minutes>=223.25 277 64 no (0.76895307 0.23104693)
18) total_eve_minutes< 242.35 210 18 no (0.91428571 0.08571429) *
19) total_eve_minutes>=242.35 67 21 yes (0.31343284 0.68656716)
38) total_night_minutes< 174.2 17 4 no (0.76470588 0.23529412) *
39) total_night_minutes>=174.2 50 8 yes (0.16000000 0.84000000) *
5) number_customer_service_calls>=3.5 164 80 yes (0.48780488 0.51219512)
10) total_day_minutes>=160.2 95 22 no (0.76842105 0.23157895)
20) state=AL,AZ,CA,CO,DC,DE,FL,HI,KS,KY,MA,MD,ME,MI,NC,ND,NE,NH,NM,OK,OR,SC,TN,VA,VT,WY 56 2 no (0.96428571 0.03571429) *
21) state=AK,AR,CT,GA,IA,ID,MN,MO,NJ,NV,NY,OH,RI,TX,UT,WA,WV 39 19 yes (0.48717949 0.51282051)
42) total_day_minutes>=182.3 21 5 no (0.76190476 0.23809524) *
43) total_day_minutes< 182.3 18 3 yes (0.16666667 0.83333333) *
11) total_day_minutes< 160.2 69 7 yes (0.10144928 0.89855072) *
3) total_day_minutes>=263.55 140 30 yes (0.21428571 0.78571429)
6) total_eve_minutes< 167.3 29 7 no (0.75862069 0.24137931)
12) state=AK,AR,AZ,CO,CT,FL,HI,IN,KS,LA,MD,ND,NM,NY,OH,UT,WA,WV 21 0 no (1.00000000 0.00000000) *
13) state=IA,MA,MN,PA,SD,TX,WI 8 1 yes (0.12500000 0.87500000) *
7) total_eve_minutes>=167.3 111 8 yes (0.07207207 0.92792793) *
R> ore.load(name=paste(datastorePrefix,"yes","yes",sep="_"))
[1] "mod"
R> mod
n= 92
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 92 36 no (0.60869565 0.39130435)
2) total_intl_minutes< 13.1 71 15 no (0.78873239 0.21126761)
4) total_intl_calls>=2.5 60 4 no (0.93333333 0.06666667)
8) state=AK,AR,AZ,CO,CT,DC,DE,FL,GA,HI,ID,IL,IN,KS,MD,MI,MO,MS,MT,NC,ND,NE,NH,NJ,OH,SC,SD,UT,VA,WA,WV,WY 53 0 no (1.00000000 0.00000000) *
9) state=ME,NM,VT,WI 7 3 yes (0.42857143 0.57142857) *
5) total_intl_calls< 2.5 11 0 yes (0.00000000 1.00000000) *
3) total_intl_minutes>=13.1 21 0 yes (0.00000000 1.00000000) *
Using the ore.rowApply Function
The ore.rowApply function invokes an R script with an ore.frame as the input data. The ore.rowApply function passes the ore.frame to the user-defined input function as the first argument to that function. The rows argument to the ore.rowApply function specifies the number of rows to pass to each invocation of the user-defined R function. The last chunk or rows may have fewer rows than the number specified. The ore.rowApply function can use data-parallel execution, in which one or more R engines perform the same R function, or task, on different partitions of data.
The ore.rowApply function returns an ore.list object or an ore.frame object.
See Also:
Example 6-11 uses the e1071 package, previously downloaded from CRAN. The example also uses the mod object, which is the a Naive Bayes model created in Example 6-8, "Using the ore.tableApply Function".
Example 6-11 does the following:
-
Loads the package
e1071. -
Pushes the
irisdata set to the database as theIRIStemporary table andore.frame. -
Creates a copy of
IRISasIRIS_PREDand adds the PRED column toIRIS_PREDto contain the predictions. -
Invokes the
ore.rowApplyfunction, passing theIRISore.frameas the data source for user-defined R function and defining the function. -
The user-defined function does the following:
-
Loads the package
e1071so that it is available to the R engine or engines running in the database. -
Converts the Species column to a factor because, although the
ore.framedefined factors, when they are loaded to the user-defined function, factors appear as character vectors. -
Invokes the
predictmethod and returns theresobject, which contains the predictions in the column added to the data set
-
-
The example pulls the model to the client R session
-
Passes
IRIS_PREDas the argumentFUN.VALUE, which specifies the structure of the object that the ore.rowApplyfunction returns. -
Specifies the number of rows to pass to each invocation of the user-defined function.
-
Displays the class of
res, and invokes thetablefunction to display the Species column and the PRED column of theresobject.
Example 6-11 Using the ore.rowApply Function
library(e1071)
IRIS <- ore.push(iris)
IRIS_PRED <- IRIS
IRIS_PRED$PRED <- "A"
res <- ore.rowApply(
IRIS,
function(dat, mod) {
library(e1071)
dat$Species <- as.factor(dat$Species)
dat$PRED <- predict(mod, newdata = dat)
dat
},
mod = ore.pull(mod),
FUN.VALUE = IRIS_PRED,
rows=10)
class(res)
table(res$Species, res$PRED)
Listing for Example 6-11
R> library(e1071)
R> IRIS <- ore.push(iris)
R> IRIS_PRED <- IRIS
R> IRIS_PRED$PRED <- "A"
R> res <- ore.rowApply(
+ IRIS ,
+ function(dat, mod) {
+ library(e1071)
+ dat$Species <- as.factor(dat$Species)
+ dat$PRED <- predict(mod, newdata = dat)
+ dat
+ },
+ mod = ore.pull(mod),
+ FUN.VALUE = IRIS_PRED,
+ rows=10)
R> class(res)
[1] "ore.frame"
attr(,"package")
[1] "OREbase"
R> table(res$Species, res$PRED)
setosa versicolor virginica
setosa 50 0 0
versicolor 0 47 3
virginica 0 3 47
Using the ore.indexApply Function
The ore.indexApply function executes the specified user-defined input function using data that is generated by the input function. It supports task-parallel execution, in which one or more R engines perform the same or different calculations, or task. The times argument to the ore.indexApply function specifies the number of times that the input function executes in the database. Any required data must be explicitly generated or loaded within the input function.
The ore.indexApply function returns an ore.list object or an ore.frame object.
Examples of the use of the ore.indexApply function are in the following topics:
See Also:
Simple Example of Using the ore.indexApply Function
Example 6-12 invokes ore.indexApply and specifies that it execute the input function five times in parallel. It displays the class of the result, which is ore.list, and then displays the result.
Example 6-12 Using the ore.indexApply Function
res <- ore.indexApply(5,
function(index) {
paste("IndexApply:",index)
},
parallel=TRUE)
class(res)
res
R> res <- ore.indexApply(5,
+ function(index) {
+ paste("IndexApply:",index)
+ },
+ parallel=TRUE)
R> class(res)
[1] "ore.list"
attr(,"package")
[1] "OREembed"
R> res
$`1`
[1] "IndexApply: 1"
$`2`
[1] "IndexApply: 2"
$`3`
[1] "IndexApply: 3"
$`4`
[1] "IndexApply: 4"
$`5`
[1] "IndexApply: 5"
Column-Parallel Use Case
Example 6-12 uses the R summary function to compute in parallel summary statistics on the first four numeric columns of the iris data set. The example combines the computations into a final result. The first argument to the ore.indexApply function is 4, which specifies the number of columns to summarize in parallel. The user-defined input function takes one argument, index, which will be a value between 1 and 4 and which specifies the column to summarize.
The example invokes the summary function on the specified column. The summary invocation returns a single row, which contains the summary statistics for the column. The example converts the result of the summary invocation into a data.frame and adds the column name to it.
The example next uses the FUN.VALUE argument to the ore.indexApply function to define the structure of the result of the function. The result is then returned as an ore.frame object with that structure.
Example 6-13 Using the ore.indexApply Function and Combining Results
res <- NULL
res <- ore.indexApply(4,
function(index) {
ss <- summary(iris[, index])
attr.names <- attr(ss, "names")
stats <- data.frame(matrix(ss, 1, length(ss)))
names(stats) <- attr.names
stats$col <- names(iris)[index]
stats
},
FUN.VALUE=data.frame(Min.=numeric(0),
"1st Qu."=numeric(0),
Median=numeric(0),
Mean=numeric(0),
"3rd Qu."=numeric(0),
Max.=numeric(0),
col=character(0)),
parallel=TRUE)
res
Listing for Example 6-13
R> res <- NULL
R> res <- ore.indexApply(4,
+ function(index) {
+ ss <- summary(iris[,index])
+ attr.names <- attr(ss,"names")
+ stats <- data.frame(matrix(ss,1,length(ss)))
+ names(stats) <- attr.names
+ stats$col <- names(iris)[index]
+ stats
+ },
+ FUN.VALUE=data.frame(Min.=numeric(0),
+ "1st Qu."=numeric(0),
+ Median=numeric(0),
+ Mean=numeric(0),
+ "3rd Qu."=numeric(0),
+ Max.=numeric(0),
+ col=character(0)),
+ parallel=TRUE)
R> res
Min. X1st.Qu. Median Mean X3rd.Qu. Max. col
1 2.0 2.8 3.00 3.057 3.3 4.4 Sepal.Width
2 4.3 5.1 5.80 5.843 6.4 7.9 Sepal.Length
3 0.1 0.3 1.30 1.199 1.8 2.5 Petal.Width
4 1.0 1.6 4.35 3.758 5.1 6.9 Petal.Length
Warning message:
ORE object has no unique key - using random order
Simulations Use Case
You can use the ore.indexApply function in simulations, which can take advantage of high-performance computing hardware like an Oracle Exadata Database Machine. Example 6-14 takes multiple samples from a random normal distribution to compare the distribution of the summary statistics. Each simulation occurs in a separate R engine in the database, in parallel, up to the degree of parallelism allowed by the database.
Example 6-14 defines variables for the sample size, the mean and standard deviations of the random numbers, and the number of simulations to perform. The example specifies num.simulations as the first argument to the ore.indexApply function. The ore.indexApply function passes num.simulations to the user-defined function as the index argument. This input function then sets the random seed based on the index so that each invocation of the input functions generates a different set of random numbers.
The input function next uses the rnorm function to produce sample.size random normal values. It invokes the summary function on the vector of random numbers, and then prepares a data.frame as the result it returns. The ore.indexApply function specifies the FUN.VALUE argument so that it returns an ore.frame that structures the combined results of the simulations. The res variable gets the ore.frame returned by the ore.indexApply function.
To get the distribution of samples, the example invokes the boxplot function on the data.frame that is the result of using the ore.pull function to bring selected columns from res to the client.
Example 6-14 Using the ore.indexApply Function in a Simulation
res <- NULL
sample.size = 1000
mean.val = 100
std.dev.val = 10
num.simulations = 1000
res <- ore.indexApply(num.simulations,
function(index, sample.size=1000, mean=0, std.dev=1) {
set.seed(index)
x <- rnorm(sample.size, mean, std.dev)
ss <- summary(x)
attr.names <- attr(ss,"names")
stats <- data.frame(matrix(ss,1,length(ss)))
names(stats) <- attr.names
stats$index <- index
stats
},
FUN.VALUE=data.frame(Min.=numeric(0),
"1st Qu."=numeric(0),
Median=numeric(0),
Mean=numeric(0),
"3rd Qu."=numeric(0),
Max.=numeric(0),
index=numeric(0)),
parallel=TRUE,
sample.size=sample.size,
mean=mean.val, std.dev=std.dev.val)
options("ore.warn.order" = FALSE)
head(res, 3)
tail(res, 3)
boxplot(ore.pull(res[,1:6]),
main=sprintf("Boxplot of %d rnorm samples size %d, mean=%d, sd=%d",
num.simulations, sample.size, mean.val, std.dev.val))
Listing for Example 6-14
R> res <- ore.indexApply(num.simulations,
+ function(index, sample.size=1000, mean=0, std.dev=1) {
+ set.seed(index)
+ x <- rnorm(sample.size, mean, std.dev)
+ ss <- summary(x)
+ attr.names <- attr(ss,"names")
+ stats <- data.frame(matrix(ss,1,length(ss)))
+ names(stats) <- attr.names
+ stats$index <- index
+ stats
+ },
+ FUN.VALUE=data.frame(Min.=numeric(0),
+ "1st Qu."=numeric(0),
+ Median=numeric(0),
+ Mean=numeric(0),
+ "3rd Qu."=numeric(0),
+ Max.=numeric(0),
+ index=numeric(0)),
+ parallel=TRUE,
+ sample.size=sample.size,
+ mean=mean.val, std.dev=std.dev.val)
R> options("ore.warn.order" = FALSE)
R> head(res, 3)
Min. X1st.Qu. Median Mean X3rd.Qu. Max. index
1 67.56 93.11 99.42 99.30 105.8 128.0 847
2 67.73 94.19 99.86 100.10 106.3 130.7 258
3 65.58 93.15 99.78 99.82 106.2 134.3 264
R> tail(res, 3)
Min. X1st.Qu. Median Mean X3rd.Qu. Max. index
1 65.02 93.44 100.2 100.20 106.9 134.0 5
2 71.60 93.34 99.6 99.66 106.4 131.7 4
3 69.44 93.15 100.3 100.10 106.8 135.2 3
R> boxplot(ore.pull(res[,1:6]),
+ main=sprintf("Boxplot of %d rnorm samples size %d, mean=%d, sd=%d",
+ num.simulations, sample.size, mean.val, std.dev.val))
Figure 6-2 Display of the boxplot Function in Example 6-14
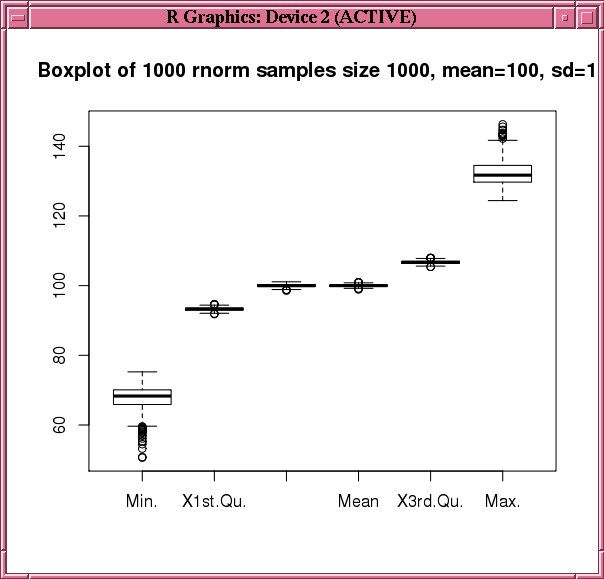
Description of "Figure 6-2 Display of the boxplot Function in Example 6-14"
Using the ore.scriptCreate and ore.scriptDrop Functions
The ore.scriptCreate function creates a script in R script repository of the Oracle database. Embedded R execution functions can use a script from the repository by specifying it with the FUN.NAME argument. Scripts in the R script repository are also available through the SQL interface for Oracle R Enterprise embedded R execution.
The ore.scriptDrop function removes the specified R script from the R script repository.
Note:
Invoking theore.scriptCreate or ore.scriptDrop function requires the RQADMIN role.Both the ore.scriptCreate and ore.scriptDrop functions return an invisible NULL value if it succeeds; if it does not succeed in creating or dropping the script, it returns an error.
Example 6-15 creates a script and stores it in the R script repository. The use-defined function in the script creates a linear model. The example pushes the iris data set to the database. It then invokes the ore.tableApply function and specifies the stored script with the FUN.NAME argument. The example then drops the script from the repository.
Example 6-15 Using the ore.scriptCreate and ore.scriptDrop Functions
ore.scriptCreate("MYLM",function(data, formula, ...) lm(formula, data, ...))
IRIS <- ore.push(iris)
ore.tableApply(IRIS[1:4], FUN.NAME = "MYLM", formula = Sepal.Length ~ .)
ore.scriptDrop("MYLM")
Listing for Example 6-15
R> ore.scriptCreate("MYLM",function(data, formula, ...) lm(formula, data, ...))
R> IRIS <- ore.push(iris)
R> ore.tableApply(IRIS[1:4], FUN.NAME = "MYLM", formula = Sepal.Length ~ .)
Call:
lm(formula = formula, data = data)
Coefficients:
(Intercept) Sepal.Width Petal.Length Petal.Width
1.8560 0.6508 0.7091 -0.5565
R> ore.scriptDrop("MYLM")
See Also:
Example 6-10SQL Interface for Embedded R Execution
The SQL interface for Oracle R Enterprise embedded R execution allows you to execute R scripts in production database applications.
The functions to use with the SQL interface must be stored in the database R repository, and referenced by name in SQL API functions. See Registering and Managing SQL Scripts for a description of how to add scripts to the repository, remove scripts from the repository, and list and use scripts in the repository.
For descriptions of the SQL functions, see About Oracle R Enterprise SQL Functions.
This section describes the SQL interface in the following topics:
Registering and Managing SQL Scripts
The R functions to use with the SQL interface for embedded R execution must be stored in the database R script repository and be referenced by name in SQL API functions. For security purposes, you must first register the R script under some system unique name and then use new name instead of the actual script in call to SQL interface table functions.
The administrative functions sys.rqScriptCreate and sys.rqScriptDrop create and drop scripts. The sys.rq_scripts view allows you to list and use scripts that were created.
Script creation or deletion requires the RQADMIN role described in "Security Considerations for Scripts".
When using the sys.rqScriptCreate function, you must specify a corresponding R closure of the function string.
Example 6-16 demonstrates registering an R script and using it.
About Oracle R Enterprise SQL Functions
Invoking an Oracle R Enterprise SQL function results in one or more R engines being started at the database depending on database parallelism settings. To enable execution of an R script in the database, Oracle R Enterprise provides SQL variants of the ore.doEval, ore.tableApply, ore.groupApply, and ore.rowApply functions. Those R functions are described in "R Interface for Embedded R Execution".
The SQL functions for embedded R execution are:
-
rqTableEval -
rqEval -
rqRowEval -
rqGroupEval
The rqGroupEval function requires additional SQL specification and is provided here as a virtual function that partitions the data according to the values of a specified column and invokes the R script on each partition. For more information, see "Using the rqGroupEval Function".
You can also use these functions with objects in a datastore, as described in "Datastore Management in SQL".
The rqEval, rqTableEval, rqGroupEval, and rqRowEval) functions have similar syntax:
rq*Eval(
cursor(select * from table-1),
cursor(select * from table-2),
'select <column list> from table-3 t',
<grouping col-name from table-1 or num_rows>,
<R closure name of registered-R-code>
)
The following are the components of the SQL function:
-
The first cursor is the input cursor: Input is passed as a whole table, group, or N rows at a time to the R closure described in the fourth argument.
The
rqEvalfunction does not have this cursor argument. -
The second cursor is the parameters cursor: One row of scalar values (string, numeric, or both) can be passed; for example, the name of the model and several numeric scalar values for model setting.
-
The query specifies the output table definition; output can be 'SELECT statement', 'XML', or 'PNG'.
-
grouping col-nameapplies torqGroupEval; it provides the name of the grouping column. -
num_rowsapplies torqRowEval; it provides the number of rows to provide to the functions at one time. -
<R closure name of registered-R-code> is a registered version of the R function to execute. See "Registering and Managing SQL Scripts" for details.
The return values for all of the SQL functions specify one of these values:
-
A table signature that is specified in a SELECT statement, which returns results as a table from the
rqfunction. -
XML, returned as a CLOB that contains both structured and graph images in an XML string. The structured components are provided first, followed by the base 64 encoding of the
pngrepresentation of the image. -
PNG, returned as a BLOB that contains graph images in
pngformat.
The rqEval, rqTableEval, rqGroupEval, and rqRowEval functions must specify an R script by the name that is stored in the R script repository. The sys.rq_scripts view provides a list of registered scripts.
The following examples illustrate using these functions:
-
This example uses all rows from the table
fishas input to the R function that takes no other arguments and producesoutputthat contains all input data plus the ROWSUM of values.Note that
paramargument to the R function is optional.begin sys.rqScriptCreate('tmrqfun2', 'function(x, param) { dat <- data.frame(x, stringsAsFactors=F) cbind(dat, ROWSUM = apply(dat,1,sum)+10) }'); end; / select * from table(rqTableEval( cursor(select * from fish), NULL, 'select t.*, 1 rowsum from fish t', 'tmrqfun2' )); begin sys.rqScriptDrop('tmrqfun2'); end; / -
This example illustrates passing n=1 (4th parameter) row at a time from the table
fishto the R function. No parameters are required by the function. The function generates ROWSUM which is added as an extra column tofishin the output.begin sys.rqScriptCreate('tmrqfun2', 'function(x, param) { dat <- data.frame(x, stringsAsFactors=F) cbind(dat, ROWSUM = apply(dat,1,sum)+10) }'); end; / select * from table(rqRowEval( cursor(select * from fish), NULL, 'select t.*, 1 rowsum from fish t', 1, 'tmrqfun2' )); begin sys.rqScriptDrop('tmrqfun2'); end; /
Using the rqGroupEval Function
The rqGroupEval function invokes an R script on data that is partitioned by a grouping column. The rqGroupEval function requires the creation of the following two PL/SQL objects:
-
A PL/SQL package that specifies the types of the result to return.
-
A function that takes the return value of the package and uses the return value with
PIPELINED_PARALLEL_ENABLEset to indicate the column on which to partition data.
Suppose that ONTIME_S is a table that stores information about arrival of airplanes. The data cursor uses all data, but you could also define cursors that use some columns using PL/SQL records. Then you must define as many PL/SQL table functions as the number of grouping columns that you are interested in using for a particular data cursor.
CREATE PACKAGE ontimePkg AS TYPE cur IS REF CURSOR RETURN ontime_s%ROWTYPE; END ontimePkg; / CREATE FUNCTION ontimeGroupEval( inp_cur ontimePkg.cur, par_cur SYS_REFCURSOR, out_qry VARCHAR2, grp_col VARCHAR2, exp_txt CLOB) RETURN SYS.AnyDataSet PIPELINED PARALLEL_ENABLE (PARTITION inp_cur BY HASH (month)) CLUSTER inp_cur BY (month) USING rqGroupEvalImpl; /
At this time, only one grouping column is supported. If you have multiple columns, then combine the columns into one column and use the new column as a grouping column. The PARALLEL_ENABLE clause is optional but the CLUSTER BY clause is not.
Using SQL Functions and Objects in a Datastore
The SQL functions for embedded R execution allow you to use in a parameter cursor a serialized R object saved in a datastore. You can specify the association of object and datastore names of the serialized R objects with the R function parameter names in that parameter cursor.
Example 6-17 demonstrates using the rqTableEval function and specifying a datastore in a cursor. The example uses a datastore named ontime_model and gets the lm.mod model object from the datastore. The example uses the model in SQL for scoring using embedded R execution.
Example 6-17 Using rqTableEval and Specifying a Datastore
begin
sys.rqScriptCreate('tmrqmodelscore',
'function(dat, in.dsname, in.objname) {
ore.load(name=in.dsname, list=in.objname)
mod <- get(in.objname)
prd <- predict(mod, newdata=dat)
prd[as.integer(rownames(prd))] <- prd
res <- cbind(dat, PRED = prd)
res
}');
end;
/ -- score model
select * from table(rqTableEval(
cursor(select ARRDELAY, DISTANCE, DEPDELAY from ontime_s
where year = 2003 and month = 5 and dayofmonth = 2),
cursor(select 'ontime_model' as "in.dsname",
'lm.mod' as "in.objname", 1 as "ore.connect" from dual),
'select ARRDELAY, DISTANCE, DEPDELAY, 1 PRED from ontime_s',
'tmrqmodelscore'))
order by 1, 2, 3;
Datastore Management in SQL
Oracle R Enterprise provides basic management for datastores in SQL. Basic datastore management includes show, search, and drop. The following functions and views are provided:
-
rqDropDataStoredeletes a datastore and all of the objects in the datastore.Syntax:
rqDropDataStore('<ds_name>'), where<ds_name>is the name of the datastore to delete.The following example deletes the datastore ds_model from current user schema:
rqDropDataStore('ds_model') -
rquser_DataStoreListis a view containing datastore-level information for all datastores in the current user schema. The information consists of datastore name, number of objects, size, creation date, and description.These examples illustrate using the view:
select * from rquser_DataStoreList; select dsname, nobj, dssize from rquser_datastorelist where dsname = 'ds_1';
-
rquser_DataStoreContentsis a view containing object-level information about all datastores in the current user schema. The information consists of object name, size, class, length, number of rows and columns.This example lists the datastore contents for datastore
ds_1:select * from rquser_DataStoreContents where dsname = 'ds_1';