| Oracle® Health Sciences Translational Research Center User's Guide Release 3.1.0.3 E66623-05 |
|
|
PDF · Mobi · ePub |
| Oracle® Health Sciences Translational Research Center User's Guide Release 3.1.0.3 E66623-05 |
|
|
PDF · Mobi · ePub |
This chapter contains the following topics:
In CE 3.1, a set of cohort viewers are supplied that enables the user to view patients or subjects in a variety of formats. You can view patient or subject details as a tabular list, or in a timeline view. You can drill into each single patient or subject details and see them all in one page. Furthermore, if Omics Data Bank model is licensed, you can look at patients or subjects genomic data in a circular genome viewer (using Visquick) or export patients or subjects data into formats acceptable by the Integrative Genome Viewer. The following sections describe the viewer options available in more detail.
Once you run a query and can see the patient or subject count, you may want to review the data for those specific patients or subjects. To view a list, select the next tab to the right, the Cohort Viewer tab. This tab displays one row of data for each patient or subject represented in a query count. To the left of the Cohort List main window, there is a separate pane where you can select and filter the data you want to view for your Cohort List.
By default, this tab displays the option to list patients or subjects for the current active query, that is, the query currently loaded in the Cohort Query tab. To view this list, select Submit.
Alternatively, you can to view a patient or subject list for a query from the library, saved patient or subject list, or from the current omics query. You can specify one or more patient or subject (study) ID's on an ad-hoc basis if you have specific patients or subjects you want to examine.
Select the arrow to the left of Data to display the list of check boxes for data topics. These topics reflect the query selection criteria from the Clinical Information category. To view the data from the selected topics, perform the following steps:
Select one or more of the check boxes for the data you want to display in your list.
Select Submit.
The system reruns the query and adds relevant patient or subject data for each patient or subject listed.
To remove data, clear the appropriate boxes and select Submit.
To export the list to a Microsoft Excel sheet, click Export. To display the date in the format dd/mm/yyyy, use the formatting option in Microsoft Excel.
Note:
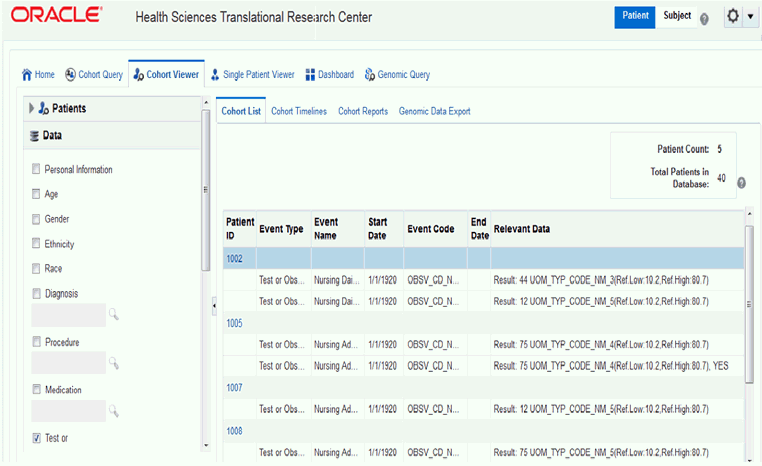
A patient or subject may have more than one row of data for a particular data category (a patient or subject can have more than one procedure or medication). As a result, the patient or subject details display may show multiple rows or records for each patient or subject. Selecting several data categories to display may significantly limit your ability to review multiple patients or subject at the same time. This feature intends to help you visualize how the query criteria are manifested in the actual clinical information.Cohort List supports displaying Reference Range values (if available) along with numeric results of Observation.
If the selected observation event data has the reference high and reference low range values along with numeric results, then the reference range values are displayed along with the numeric result values in the Relevant Data column of the cohort list.
Figure 5-3 Cohort List with Reference Range Values for Numeric Results of Observation
To view patient or subject data in a list, CE has a Timelines Viewer which provides a method to view data for a small subset of patients in a more visual way. You can select data topics such as Diagnoses, Procedures, Medications to view the details and the system displays when specific activities occurred, in context with each other. Or, the view displays that a procedure was performed before or after a particular diagnosis was identified or a particular medication was taken by the patient or subject.
The first step is to select the patients or subjects. This is similar to how you select patients or subjects for the Cohort List. The selection for the viewer is done by selecting patients or subjects either from the current active query which displays by default, from a query in the library, or by entering the Patient or Subject ID number for the patients or subjects you want to examine.
Figure 5-4 Selecting Patients or Subjects
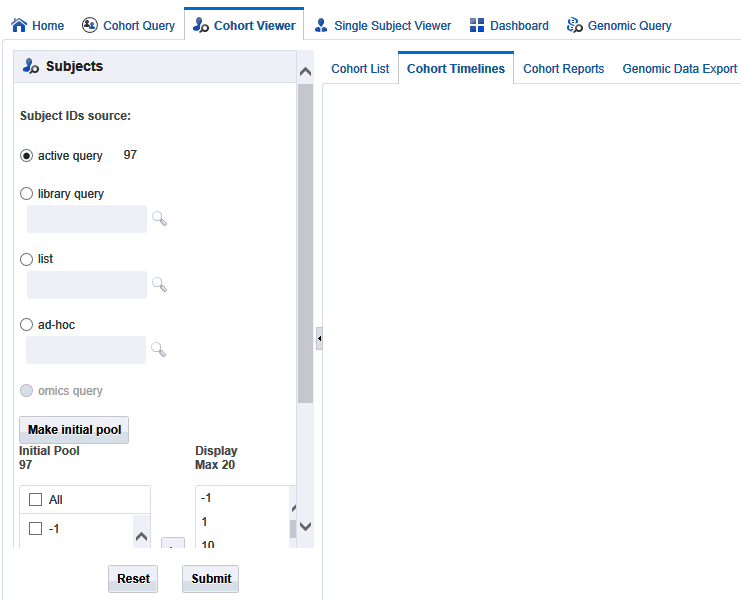
To select patients or subjects to view perform the following steps:
The upper portion of the Patients or Subjects filter is where you specify patients or subjects from the active query, or library query, or particular Patient or Subject ID. Once you have chosen where the patients or subjects come from, you select the Make Initial Pool button (item 1 in Figure 5-5), and the patients or subjects to be added to the left hand list below the button. The last step is to select the patients or subjects to add to the Initial Pool.
The next step is to select up to 20 patients or subjects and move them from the Initial Pool to the Display List. This is done by clicking the right hand arrow (Item 2 in Figure 5-5). The Display List is what the system will reference for displaying the data in the timelines view.
Click Submit to view the corresponding data. However, you may select Clinical data for these Patients or Subjects, as outlined in the Patient or Subject Pool section.
Once you have identified patients or subjects for the Display List, you then select the clinical information to be displayed in the viewer.
At the bottom of the left hand panel, click the arrow next to Data. The system displays a list of data topics.
Select the box for each data topic you want to view. Then select the magnifying glass icon for each selected topic to search or specify the particular criteria. A popup search window is displayed.
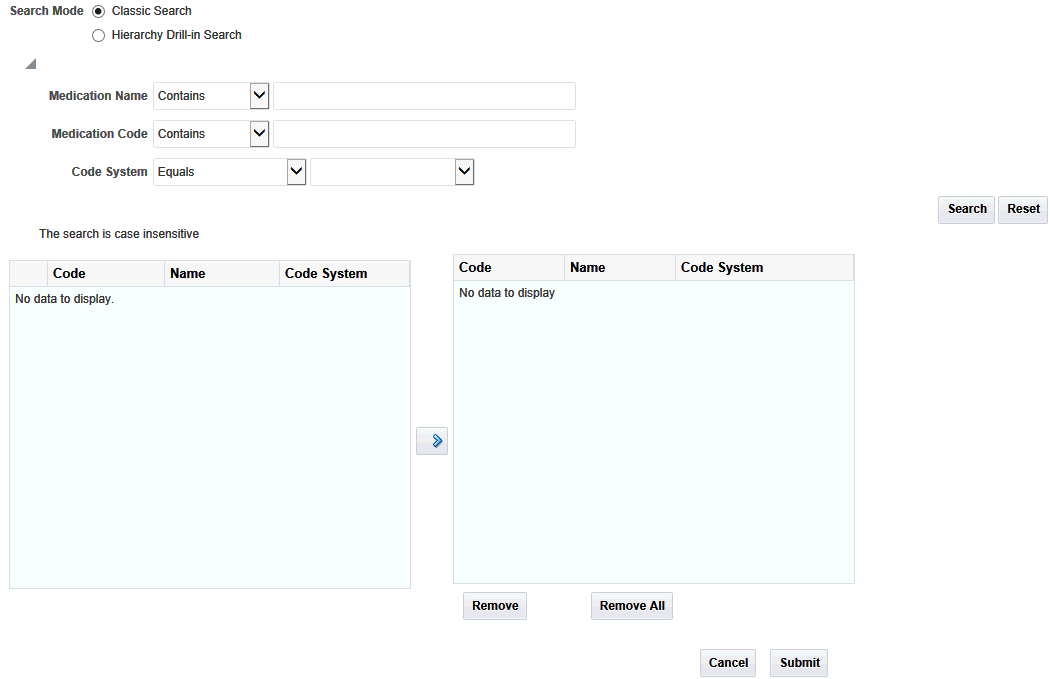
For each topic, enter the Name, Code, or Code System.
Click Search.
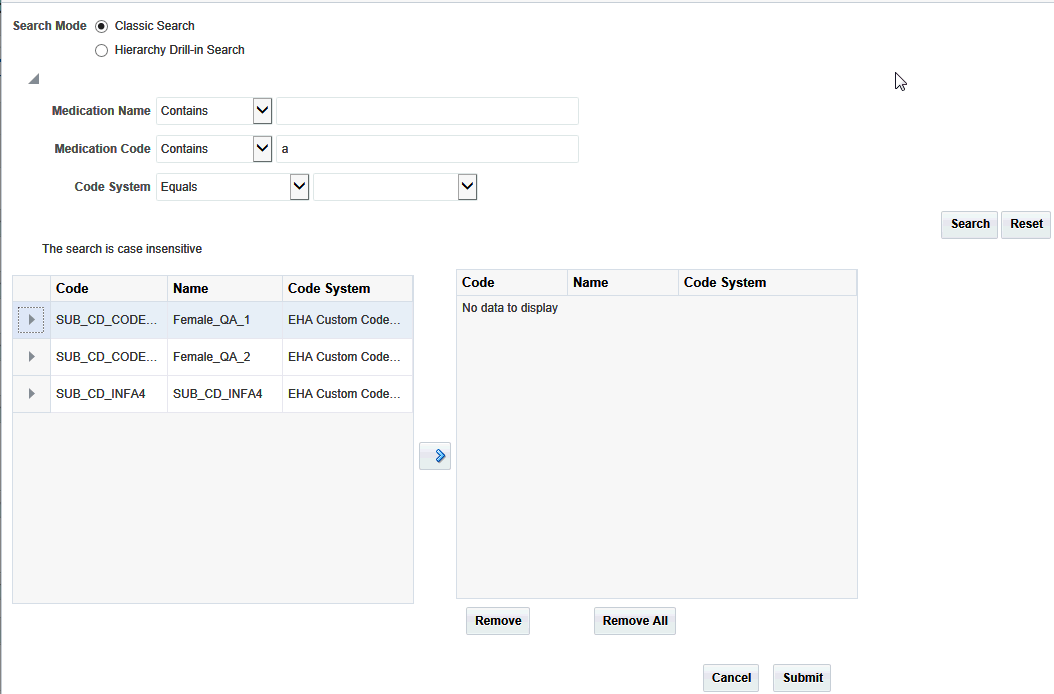
Select one or more items and use the right hand arrow to move them to the right hand box.
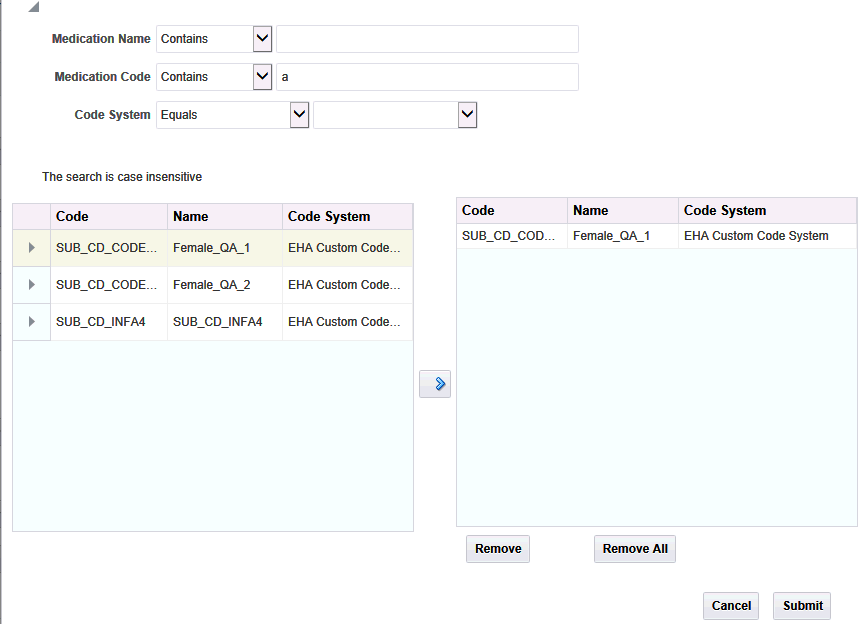
Click Submit.
Each data topic has a distinct color associated with it, because the data will be aligned in a timelines sequence, and the color serve as a visual separator between different data for the same patient or subject. CE has assigned default colors, but you can select any one of the drop down arrows to change that topic's color to suit your needs.
Once you have selected and specified the data you want to view, select Submit at the bottom of the list. The system displays a linear view of the selected data in the main window of the timelines viewer.
The display is subdivided into two sections, with a narrow vertical divider. The left side has attributes about the selected patients and the right side shows the colored layers of the selected clinical data, in a timeline view. You can move the divider left or right by hovering the cursor on the white vertical line. Select and hold down your mouse and then drag the divider to the desired position.
Also, you can hide the display of the patient or subject and data selection pane as well as the left hand side of the timelines view, by clicking the small left hand arrow on the right hand side of the respective area.
In Figure 5-7, the first column is the patient ID. Clicking it will navigate to the single patient viewer.
Note:
If you are using Mozilla Firefox, click the patient ID. For Microsoft Internet Explorer and Google Chrome, double-click the patient ID.Using the option View same Events, you can display the events in one of the following modes:
This is the default mode of displaying data on the Timelines tab.
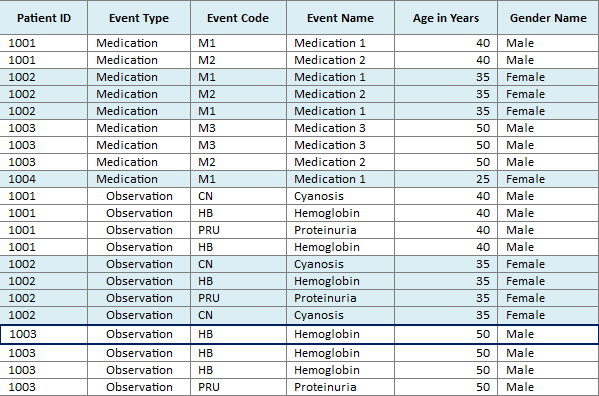
In this mode, all instances of a repeating event are displayed on the same line and separate events are displayed on separate lines. The events are considered to be repeating on the basis of Patient, Event Type and Event Code. For example, if a patient P1 has taken the medication M1 from 01-March to 31-March and then from 15-August to 30-September, then the Medication Event M1 is considered to be repeating for the patient and this mode displays the two occurrences of the medication event in the same line in the timeline.
If there is an overlap of events, for example, the start date of an event is the same as or earlier than the end date of the preceding occurrence of the event, then such overlapping occurrences are displayed in separate lines in the timeline. However, the left hand table displays a single row corresponding to all occurrences of the event.
In the single line mode, the tabular display on the left side uses alternating background color to visually delineate data of different patients or subjects. All of the rows corresponding to a patient or subject, for a particular Event Type, have one background color and the next set of rows are in a different color.
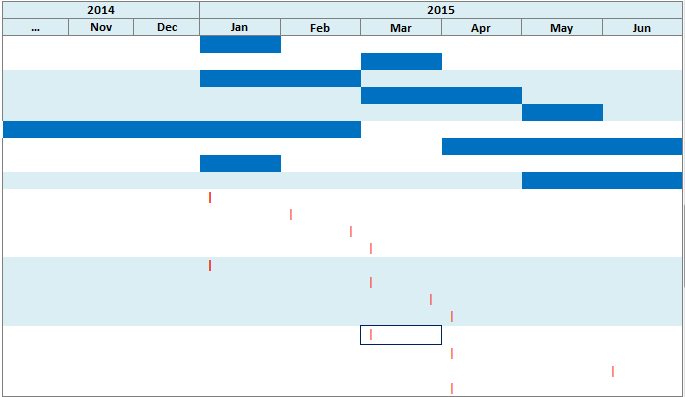
In this mode, all of the instances of events are displayed on different lines, even if the events are repeating. For example, the two occurrences of the medication event mentioned above are displayed in separate lines in this mode.
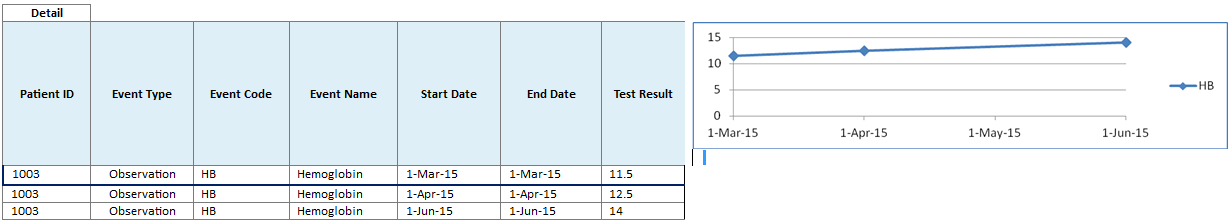
Following are some details about timelines:
If an event has both Start Date and End Date values available, then the timeline uses the Event Code to mark the start of an event.
A tooltip on the timeline provides additional information about the event.
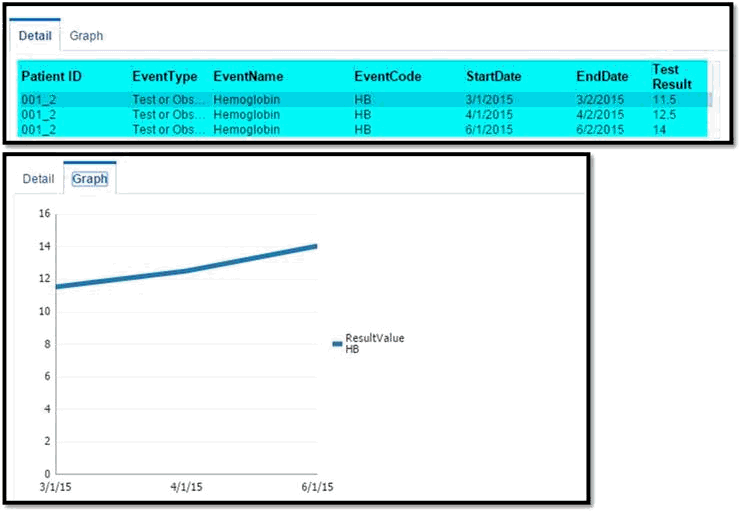
Upon selecting an event in the timeline, the Detail table below the timeline lists all the occurrences of the event and provides additional information such as the start and the end date of the occurrences.
Select only one Timeline Event at a time to render the corresponding Detail Table. If you select multiple Timeline Events, then the UI is likely to stop responding and may become unusable.
For Test or Observation events with numeric result values, the Detail table displays the results of all the occurrences of the event. If two or more occurrences of Test or Observation event have numeric results available, then the Graph tab displays a line graph for those numeric result values.
Since the Test or Observation event has only one date event, the user interface uses dummy End Date values that are 1 day later than the corresponding Start Date values. This is to support proper rendering of graph. The Detail table also displays these dummy End Date values.
Events are not displayed on the left hand table or on the timeline in the following conditions:
When an event's Start Date is null.
When an event has only one Date attribute (for example, Observation has only Observation Date) and the attribute's value is null.
If an event's End Date is null, then the System Date is used for displaying the event.
Above the data display, there are two functional controls:
The Time Scale option enables you to choose a specific date range for viewing the data. The default auto setting displays the data without a specific time reference.
The Align Data by Patient or Subject Event tab enables you to designate a particular piece of clinical data to serve as the anchor, around which the remaining data is adjusted in the viewer. For example, if you choose a particular procedure as the anchor for alignment, medications or diagnostic tests be adjusted in the display to show how long before (or after) the anchor they were administered.
When you select a particular diagnosis or medication code to align patients, all the event codes at different hierarchical nodes are considered for the selected event code.
Using the Include criteria used in query check box, you can view the data used to define the inclusion criteria of the currently active query. The criteria defined in the cohort query will be displayed in the data section of the cohort timeline.
If the inclusion criteria of the active query is not based on any data topics supported on Cohort Timeline, a message is displayed to select at least one of the data elements supported by Cohort Timeline.
Note:
The data topic Clinical Encounter can be selected only in conjunction with other data elements that are linked to Clinical Encounter, for example Diagnosis, Procedure and so on.For example, in the Figure 5-10, Diagnosis has been defined as inclusion criteria in the cohort query screen. Once you select it, the corresponding diagnosis criteria is displayed. Then click Submit to view the corresponding timeline events.
This section lets you view various cohort reports.
Note:
In the Active Query mode, if no filters have been selected in Cohort Query, then the reports display data of only the first 10,000 patients or subjects. This limit is configurable and can be changed using the DEFAULT_ACTIVE_QUERY_LIMIT property in the TRC.properties file.However, irrespective of the patient or subject selection option, if the count exceeds the value specified by the GENOMIC.REPORTS_MAX_PATIENT_COUNT option of TRC.properties file, a warning message is displayed. You can either continue to plot with a large number of patient or subjects (might impact performance) or change the selected cohort to a smaller number of patients or subject.
You can view demographic reports for a patient list. The queries for these reports can be based on patient list in the cohort query, the library, patient list, ad-hoc query or omics. Perform the following steps to view demographic reports:
Navigate to Cohort Viewer > Cohort Reports tab.
Expand the Patients node on the left.
Select the Patient IDs Source.
Expand the Demographic Reports tab.
Select the check boxes for the data you want displayed in your list.
Select Submit.
The system reruns the query and adds relevant patient or subject data for each patient or subject listed. Depending on you selection, reports age, gender, race and/or age and gender will be displayed. The data will be displayed as pie charts or bar graphs depending on your settings.
If any missing or unknown data is present, a check box is provided at the bottom of each graph to include missing or unknown data as shown in Figure 5-13.
For the Age, Gender and Race graphs, if the check box is selected, the graph is refreshed with the unknown data. Here unknown data refers to age, gender or race details that are not defined for a particular patient.
For the Age and Gender graph, the unknown age for different genders is combined into one group and the known age is in other group. Both groups are shown in the graph.
You can view clinical reports for a patient list. The queries for these reports can be based on patient list in the cohort query, the library, patient list, ad-hoc query or omics. Perform the following steps to view a clinical reports:
Navigate to Cohort Viewer > Cohort Reports tab.
Expand the Patients node on the left.
Select the Patient IDs Source.
Expand the Clinical Reports tab.
Select the Specimen Available check box.
Select Submit.
The system reruns the query and adds relevant patient or subject data for each patient or subject listed. Depending on you selection, reports age, gender, race and (or) age and gender will be displayed. The data will be displayed as pie charts or bar graphs depending on your settings.
If any missing or unknown data is present, a check box is provided at the bottom of each graph to include missing or unknown data as shown in Figure 5-15.
If Include Unknown Specimen and Label With or Include Unknown Anatomical site and Label With are selected, the graph is refreshed with the unknown data. Here unknown data refers to the specimen and anatomical site data that is not defined for a particular patient.
The following changes have been added to Genomic Reports in TRC 3.1:
An additional option to Include All Specimen is provided in all reports. This enables you to toggle while taking into account specimens, without genomic data, when generating reports.
Genomic Reports data is now filtered by Assembly; with a filter drop down provided for Assembly version.
A new node has been added to display a report on genomic data presence.
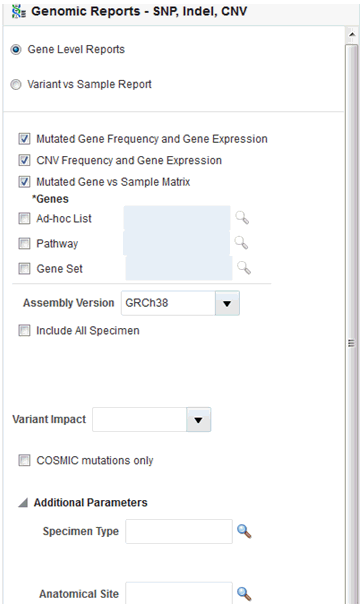
The Genomic Report—SNP, Indel, CNV—now has two reporting options, for Gene Level Reports and Variant vs. Sample Reports.
Mutated Gene vs. Sample Matrix has additional variant filters; a multi-select drop-down list Variant impact filters, and a check box to limit to Comic mutations only.
Figure 5-16 SNA, Indel, CNV Gene-Level Filter Panel
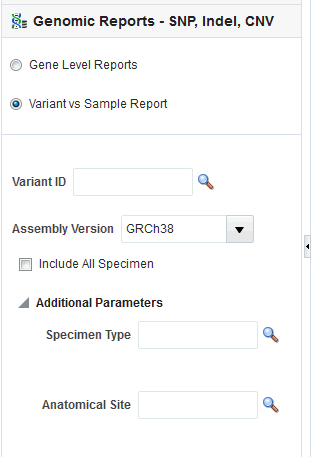
The Variant vs. Sample Report provides the option of reporting the presence of variants in specimen samples by a user input of Variant IDs.
Figure 5-17 SNA, Indel, CNV Variant vs Sample Filter Panel
The Genomic Reports - Data presence lets you view a breakup of the patient cohort as follows:
Patients in the cohort with specimens collected that have accessible genomic data
Patients in the cohort with specimens collected but no genomic data
Patients in the cohort with no specimens collected
You can view the Data Presence plot as Pie Chart, Horizontal Bars, Vertical Bars, and Table. The report also provides an option to view the distribution of present genomic data along the following available genomic data types:
Sequencing
RNA-seq
Gene Expression
Copy Number Variation
Perform the following steps to view a Genomic data presence report:
Expand the Genomic Reports - Data Presence node in the left panel.
Select the Genomic Data Presence check box. An optional Specimen Type input field appears to allow for filtering the initial cohort on specimen type.
Click Submit to generate the report.
To change the display type after generating the report, select display option from the Display as drop-down list.
To view a breakup of present genomic data by data type, click View distribution of genomic data.
To export the graphical plot type displays as a PNG image, and the table type displays as a Excel spreadsheet, click Export.
Genomic Reports - SNP, Indel and CNV display the available SNP Indel genomic reports based on the selected cohort of patients or subjects, if in subject context.
First, you must select the source of patient or subject identifiers as shown in Figure 5-21. The source of patients or subjects is same for all cohort viewers and consists of one of the following option:
active query from Cohort Query interface
saved query from a query library
saved list of identifiers
ad-hoc list of identifiers
list of patient or subject IDs based on a query performed through the Genomic Query tab
Figure 5-21 Select Source of Patient or Subject Identifiers
Next, you must show the Mutated Gene Frequence and Gene Expression report under Genomic Reports - SNP, Indel CNV category as shown in Figure 5-22. You can also opt to add additional parameters such Specimen Type, Anatomical Site, Assembly Version which will only consider results linked to the selected categories. Also, the check box Include All Specimen which when unchecked (this is the default value) will return a result which has only genomic data. If this option is selected, the result will include specimen without genomic data. Once you click Submit, a histogram report will show the percentage of samples for the relevant cohort which have sequence variants information within the selected genes.
You can display the results as horizontal bars, vertical bars, or as a table. You can also export the results into pdf if bars are exported or into Excel if table is exported.
Note:
When opening the Excel file, you may receive a warning from Excel stating that the file is in a different format than specified by the file extension. This warning can be safely ignored. For more information, refer tohttp://docs.oracle.com/cd/E23943_01/web.1111/b31973/af_table.htm#autoId34.On clicking the histogram that is displayed for the different selected genes, a popup is displayed to either get the Gene Expression plot or the Variant Viewer to display details of the variant.
Selecting Show Gene Expression link in the popup, displays the Gene Expression details for Single Channel or Dual Channel. In the Single Channel Expression, you are required to provide hybridization details to get the information. If the selected cohort and genes do not have any sequencing data associated with them, the plot is not rendered and gene expression information cannot be plotted for that selection. The gene expression plot is displayed only when the specimens of the selected cohort have both sequencing and gene expression data.
When you click Show Variant Viewer plot, a plot similar to the following is rendered:
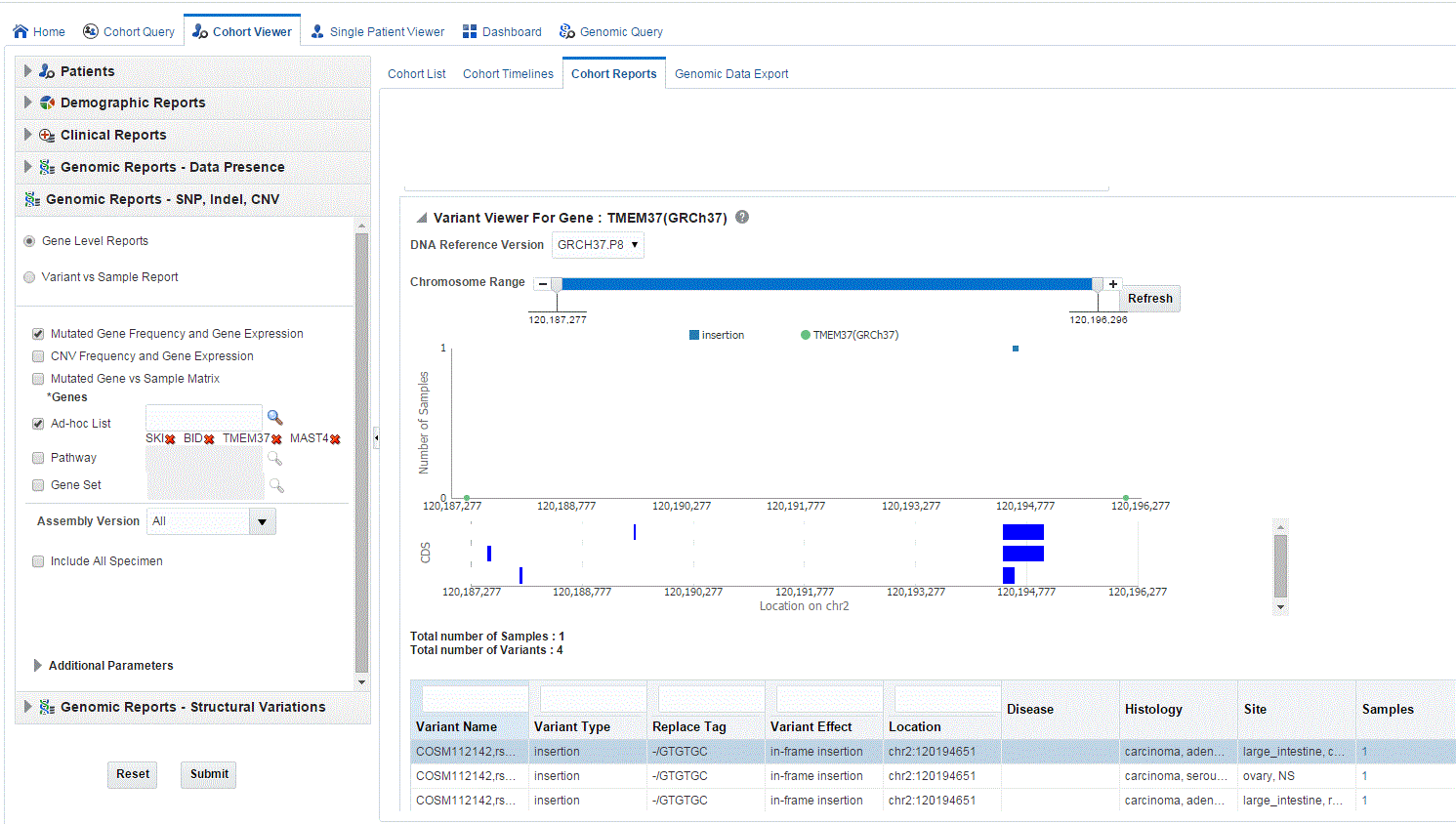
The variant viewer displays variants based on the annotation (gene coordinates) of the selected reference version. If the selected gene coordinates of the DNA Reference version change, then there can be a difference in the specimen and variant counts in the report.
This displays necessary information about all variants present in that gene in the samples belonging to the selected cohort of subjects or patients. The Mutation plot displays the total number of specimen (on y-axis) having a particular mutation (on y-axis) falling in the selected gene for belonging to a specific reference. The x-axis contain the range of gene region including the flanking region.
The CDS plot displays bars of CDS regions for each of the Ensembl Transcripts belonging to the selected gene of a selected reference version. Each row in the CDS plot represents one Transcript ID of the selected gene and reference version.
The following table represents the details of each variant like Variant name, Variant Type, Replace Tag, Variant Effect if present, Location, Disease if loaded, Histology, Site Total number of samples, Patients. The results are displayed in descending order of specimen count, such that each row contains unique variants.
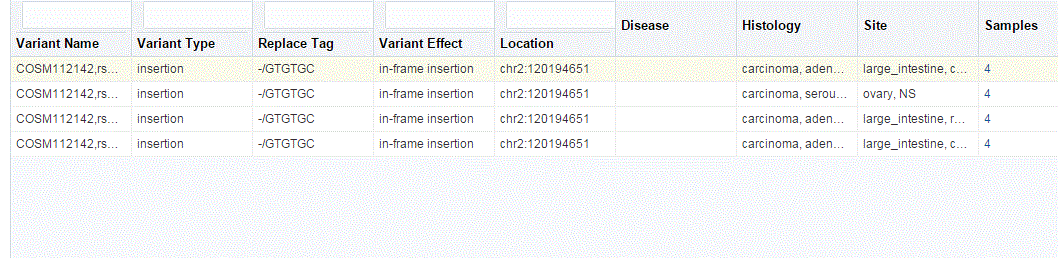
You can drill down to each mutation by clicking Number of samples present in the samples column of the table. Once clicked, a table and a pie chart are displayed below the table. The table represents the details like Sample ID, Study (only in subject context), Anatomical Site and Patient or Subject ID, which contains the selected mutation, number of samples which do not contain this mutation (labeled as Other) from the group of samples having mutations for the selected gene and selected reference version.
CNV Frequency and Gene Expression is displayed on the Genomic Reports -SNP Indel genomic reports available to the end user based on the selected cohort of patients or subjects, if in subject context.
First, you must select the source of patient or subject identifiers as shown in Figure 5-24. The source of patients or subjects is same for all cohort viewers and consists of one of the following option:
active query from Cohort Query interface
saved query from a query library
saved list of identifiers
ad-hoc list of identifiers
list of patient or subject IDs based on a query performed through the Genomic Query tab
Figure 5-24 Select Source of Patient or Subject Identifiers

Next, select to show the Copy Number Variation report under Genomic Reports - SNP, Indel CNV category as shown in Figure 5-25. You can also opt to add additional parameters such Specimen Type, Anatomical Site, Assembly Version, which will only consider results linked to the selected categories. Also, the check box Include All Specimen which when unchecked (this is the default value) will return a result which has only genomic data. If this option is selected, the result will include specimen without genomic data. After you click Submit, a histogram report will show the percentage of samples for the relevant cohort which have copy number variants information within the selected genes.
You can display the results as horizontal bars, vertical bars, or as a table. You can also export the results into pdf if bars are exported or into Excel if table is exported.
Note:
When opening the Excel file, you may receive a warning from Excel stating that the file is in a different format than specified by the file extension. This warning can be safely ignored. For more information, refer tohttp://docs.oracle.com/cd/E23943_01/web.1111/b31973/af_table.htm#autoId34.Figure 5-25 Show the Copy Number Variations
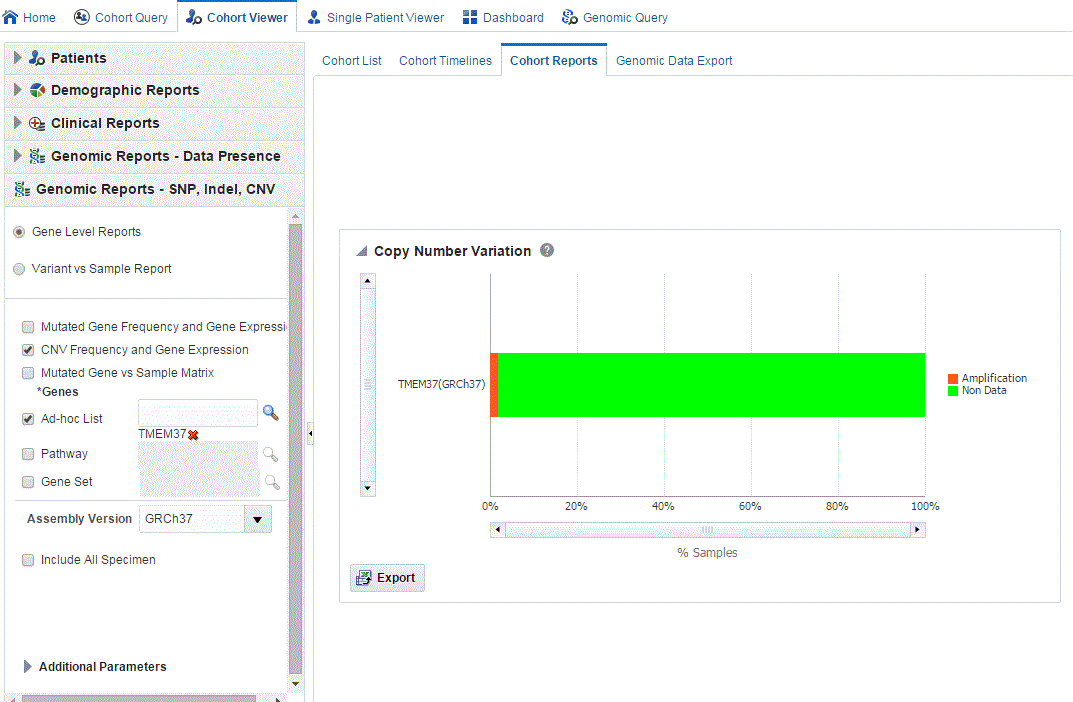
On clicking the histogram that is displayed for the different selected genes, a popup is displayed to either get the Gene Expression plot or the CNV Viewer to display details of the variant.
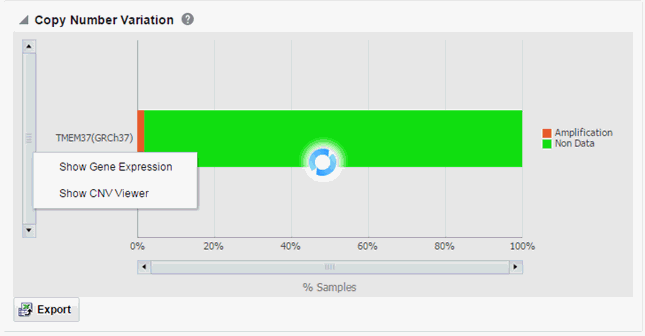
Selecting Show Gene Expression in the popup, displays the Gene Expression details for Single Channel or Dual Channel. In the Single Channel Expression you must provide hybridization details to get the information. If the selected cohort and genes do not have any sequencing data associated with them, the plot is not rendered and gene expression information cannot be plotted for that selection. The gene expression plot is displayed only when the specimens of the selected cohort have both CNV and gene expression data.
When you click Show CNV Viewer plot, then the plot similar to the following is rendered:
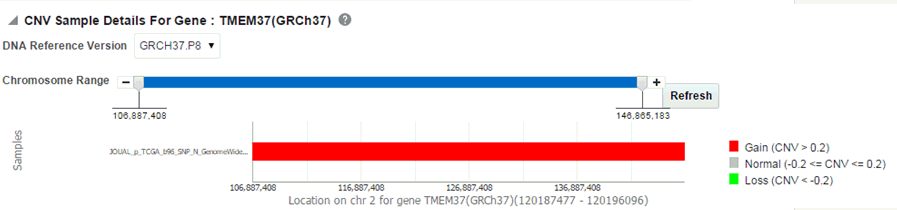
This displays necessary information about all CNV variants that are present in that gene in the samples belonging to the selected cohort of subjects/patients. The CNV plot displays each specimen (on y-axis) and its values falling in the selected gene for belonging to a specific reference.The x-axis plots the range of gene region. The plot is color coded with CNV with Gain (CNV > 0.2) is Red, Normal (-0.2<CNV<0.2) in grey and Loss (CNV < -2) in Green.
The CNV table gives the details about the CNV data like Samples, File Type, Chromosome, Anatomical Site, Start Position, End Position, CNV Value and Patient or Subject ID.

The Mutated Gene vs Sample Matrix plot displays a high level pictorial view of the presence of specific variants in genes of interest across various specimens of patients or subjects in a cohort. You can see if a particular gene of a specimen has selected mutations or no-mutations or no-data loaded. The same plot lets you view CNV data on the genes for the specimens.
The specimens are grouped together for each patient or subject and are ordered based on their collection date.
You can also view patient_id:specimen_number(specimen_vendor_number):collection_dt:no of vairants in the tooltip for each data point.
Variant selection is based on variant impact, implying that the depiction of mutations on genes for each specimen is based on the variants having selected variant impacts. For example, if you select frameshift mutation for EGFR, then the query searches for variants causing frameshift impact for EGFR gene and reports as mutation if it finds any such variants. If there is only non-variant information, then it reports as non-mutation in the plot. If there is no data, then it shows no-data in the plot.
Following are the definitions of various types of data in the plot.
Mutation: If any of the variants are present in the selected gene of a specimen, then it is reported as mutation.
No Mutation: If none of the variants are present in the selected gene of a specimen and there is non-variant information available for that gene, then it is reported as No Mutation.
No data: If there are no variants and also no non-variant information and there is no-call data for the selected gene of a specimen or there is no-data loaded for this specimen, then it will be reported as no data.
Amplification: If there is CNV data, which has seg_mean value more than zero, then it is reported as amplification.
Deletion: If there is CNV data which has seg_mean value less than zero, then it is reported as deletion.
To view this plot:
Select the source of patient or subject identifiers as shown in Figure 5-24. The source of patients or subjects is same for all cohort viewers and consists of one of the following:
Active query from Cohort Query interface
Saved query from a query library
Saved list of identifiers
Sd-hoc list of identifiers
List of patient or subject IDs based on a query performed through the Genomic Query tab
Select to show the Mutated Gene vs Sample Matrix report under Genomic Reports - SNP, Indel CNV category. Select genes as shown in Figure 5-25. Data is filtered using the Variant Impact or the cosmic mutation selected.You can also add additional parameters such Specimen Type, Anatomical Site, Assembly Version, which will only consider results linked to the selected categories. Also, the check box Include All Specimen which when unchecked (this is the default value) will return a result which has only genomic data. If this option is selected, the result will include specimen without genomic data.
After you click Submit, a report displays the percentage and details of samples for the relevant cohort which have sequence variants and copy number variants information within the selected genes.
Figure 5-26 Mutated Gene vs Sample Matrix Report
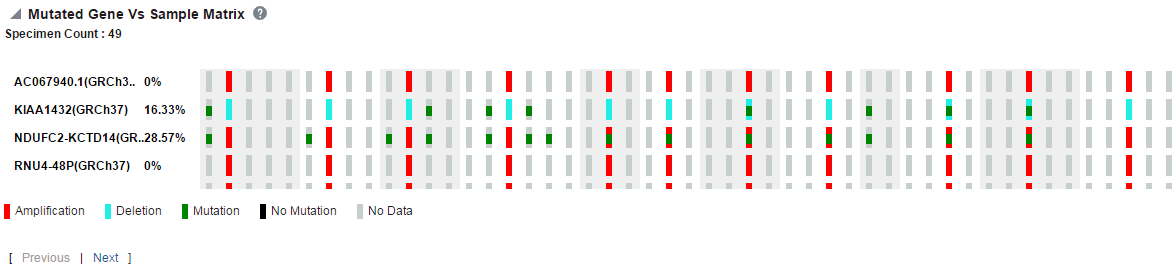
Each bar represents a specimen, which is grouped patient wise and in the order of the specimen collection date. The Sequence Variants and CNV information for each specimen is displayed. The percentage of specimens with mutation on the gene is also mentioned. Export functionality is provided at each specimen level.
For improved performance, data for only 10 genes is displayed at a time on the plot. Use the Next and Previous links to retrieve information for other set of genes. The export functionality also includes only the data of the 10 genes that is displayed in the above plot.
If the selected genes do not belong to the assembly provided for plotting, then a message stating that no CBIO data is available for selected criteria is displayed.
Note:
In rare cases, less than 10 genes might be displayed because some of the selected genes may have multiple identifiers as they are placed in different chromosomes.The data for these is clubbed together as a single gene record on the plot.
The best performance is observed for 10 genes and specimen count less than 300. As the data increases, the performance decreases linearly. Also, performance degrades with the presence of non-variant data.
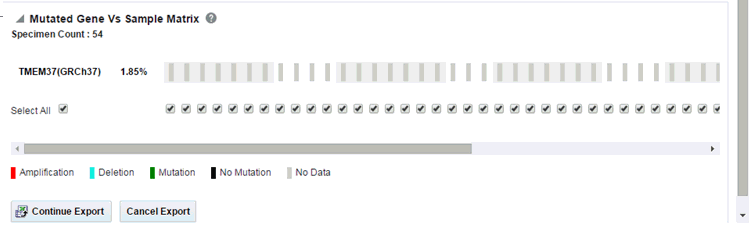
Note:
When opening the Excel file, you may receive a warning from Excel stating that the file is in a different format than specified by the file extension. This warning can be safely ignored. For more information, refer tohttp://docs.oracle.com/cd/E23943_01/web.1111/b31973/af_table.htm#autoId34.There is a limit to the number of specimens that can be seen in the gene matrix plot. This is an application level parameter called 'MAX_SPEC_REPORT' that is defined in the TRC.properties file. The default value of this parameter is 1000. If the number of specimens in the cohort used for this analysis is greater than the specified value, then the following warning message is displayed:
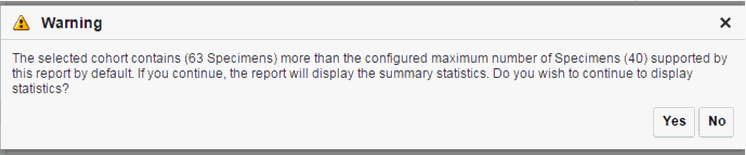
If you continue from this warning message, the report will display the summary statistics instead of the matrix plot. You may have to decrease the cohort size based on some criteria like specimen type or anatomical site or otherwise use cohort query to view the matrix plot. Alternatively, you can also change the default value of MAX_SPEC_REPORT parameter to a desired value and rebuild the plot. However, rebuilding maybe affect the performance of the plot generation.
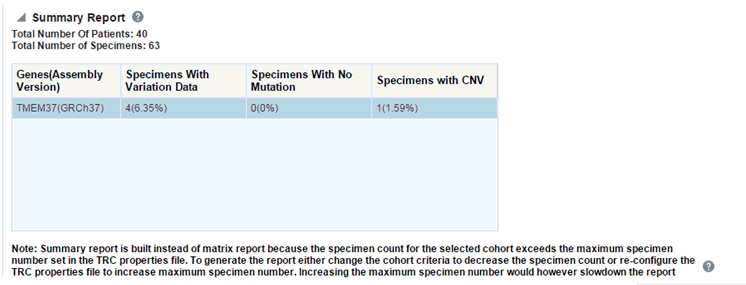
The summary report only displays the percentage of specimens with different categories of data as shown in the image above.
The Variant Level Reports option is displayed on the Genomic Reports - SNP Indel genomic reports available, based on the selected cohort of patients or subjects, if in subject context.
It provides a high level pictorial view of the presence of specific variants of interest across various specimens of patients or subjects in a cohort.
The specimens are grouped together for each patient or subject and are ordered based on their collection date. You can also view patient_id:specimen_number(specimen_vendor_number):collection_dt in the tooltip for each data point.
To generate the plot:
First, select the source of patient or subject identifiers as shown in Figure 5-24. The source of patients or subjects is same for all cohort viewers and consists of one of the following:
Active query from Cohort Query interface
Saved query from a query library
Saved list of identifiers
Sd-hoc list of identifiers
List of patient or subject IDs based on a query performed through the Genomic Query tab
Select to show the Variant Level Reports under Genomic Reports - SNP, Indel CNV category. Select variants as follows.
You can also opt to add additional parameters such Specimen Type, Anatomical Site, Assembly Version, which will only consider results linked to the selected categories. Also, the check box Include All Specimen which when unchecked (this is the default value) will return a result which has only genomic data. If this option is selected, the result will include specimen without genomic data.
After you click Submit, a report displays the percentage and details of samples for the relevant cohort, which have mutation within the selected genes.
Each bar represents a specimen, which is grouped together patient wise and in the order of the specimen collection date. The mutation information for each specimen is displayed. Each row represents one variant and the label represents reference_id, assembly version and the variant replace_tag value to maintain the uniqueness of the variant. The percentage of specimens for each mutation is also mentioned.
For improved performance, data for only 10 genes is displayed at a time on the plot. Use the Next and Previous links to retrieve information for other set of genes. The export functionality also includes only the data of the 10 genes that is displayed in the above plot.
If the selected variants do not belong to the assembly provided for plotting, then a message stating that no CBIO data is available for selected criteria is displayed.
Note:
The performance of the variant matrix report degrades if variants are encountered in the intergenic region, which will result in the query not being able to use partitioning correctly.
Note:
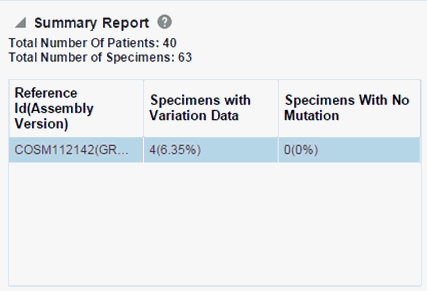
When opening the Excel file, you may receive a warning from Excel stating that the file is in a different format than specified by the file extension. This warning can be safely ignored. For more information, refer tohttp://docs.oracle.com/cd/E23943_01/web.1111/b31973/af_table.htm#autoId34.There is a limit on the number of specimens that can be seen in matrix plot. This is an application level parameter called 'MAX_SPEC_REPORT' that is defined in the TRC.properties file. The default value of this parameter is 1000. If the number of specimens in the cohort used for this analysis is greater that the specified value then the following warning message is shown below:

If you continue from this warning message, the report will display the summary statistics instead of the matrix plot. You may have to decrease cohort size based on some criteria like specimen type or anatomical site or otherwise using cohort query to view the matrix plot. Alternatively, you can also change the default value of MAX_SPEC_REPORT parameter to a desired value and rebuild the plot but the performance of the plot generation may be effected. The summary report will only display the percentage of specimens with different categories of data as shown below.
Structural Variations in Genes is one of the genomic reports available to you based on the selected cohort of patients or subjects, if it is in the subject context.
First, you must select the source of patient or subject identifiers as shown in Figure 5-28. The source of patients or subjects is same for all cohort viewers and consists of one of the following option:
active query from Cohort Query interface
saved query from a query library
saved list of identifiers
ad-hoc list of identifiers
list of patient or subject IDs based on a query performed through the Genomic Query tab
Figure 5-28 Select Source of Patient or Subject Identifiers
Next, you must show the Structural Variations (SV) in Genes report under Genomic Reports - Structural Variations category as shown in Figure 5-29. You can also add additional parameters such as specimen type, anatomical site, DNA version, which will only consider results linked to the selected categories. Once you click Submit, a histogram report shows the occurrence of structural variations involving genes sorted from the gene involved in most SVs for a given cohort, and subsequently display genes with decreasing frequency of Structural Variations. By default, top 10 genes are shown and the you can increase or decrease the number of genes displayed.
You can display the results as horizontal bars, vertical bars, or as a table. You can also export the results into pdf if bars are exported or into Excel if table is exported.
Note:
When opening the Excel file, you may receive a warning from Excel stating that the file is in a different format than specified by the file extension. This warning can be safely ignored. For more information, refer tohttp://docs.oracle.com/cd/E23943_01/web.1111/b31973/af_table.htm#autoId34.Figure 5-29 Show the Structural Variations
Structural Variations in Gene Pairs is one of the SV genomic reports available to you based on the selected cohort of patients or subjects, if it is in the subject context.
First, you must select the source of patient or subject identifiers as shown in Figure 5-30. The source of patients or subjects is same for all cohort viewers and consists of one of the following option:
active query from Cohort Query interface
saved query from a query library
saved list of identifiers
ad-hoc list of identifiers
list of patient or subject IDs based on a query performed through the Genomic Query tab
Figure 5-30 Select Source of Patient or Subject Identifiers
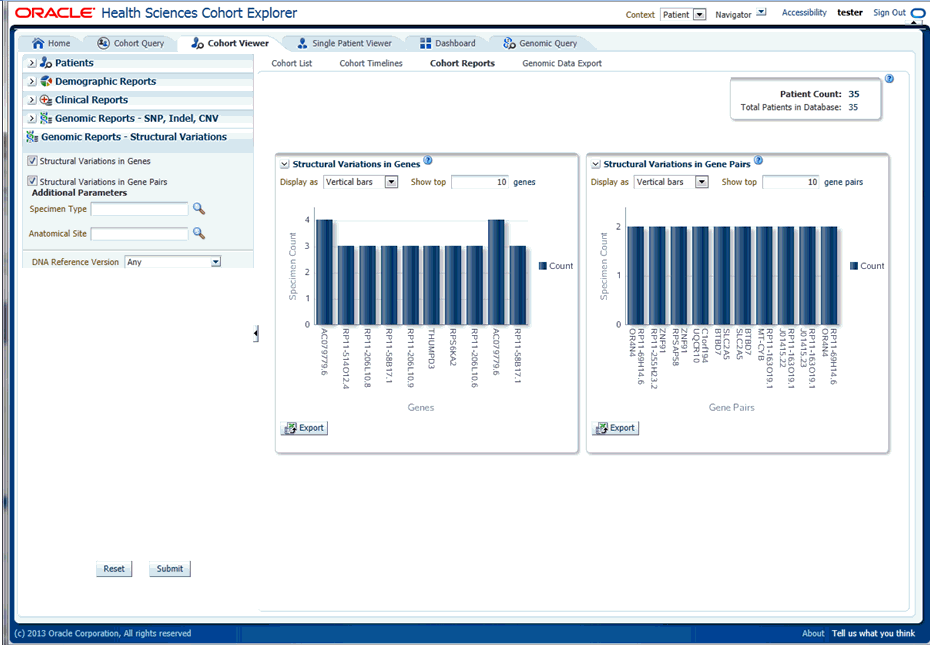
Next, you must show the Structural Variations (SV) in Gene Pairs report under Genomic Reports - Structural Variations category as shown in Figure 5-31. You can also add additional parameters such as specimen type, anatomical site, DNA version, which will only consider results linked to the selected categories. Once you click Submit, a histogram report will show the frequency of occurrence of structural variations in a cohort among gene pairs. The histogram is automatically sorted from the gene pair with most SVs as per cohort and the incidence decreases or at best stays the same for each subsequent gene pair. By default, top 10 gene pairs are shown and the user can elect to change the default number of bars shown.
You can display the results as horizontal bars, vertical bars, or as a table. You can also export the results into pdf if bars are exported or into Excel if table is exported.
Note:
When opening the Excel file, you may receive a warning from Excel stating that the file is in a different format than specified by the file extension. This warning can be safely ignored. For more information, refer tohttp://docs.oracle.com/cd/E23943_01/web.1111/b31973/af_table.htm#autoId34.Figure 5-31 Show the Structural Variations

The Genomic Data Export page is used to export the genomic data for patients or subjects filtered based on Study, Specimen type and Anatomical Site in a specific file format. Currently, exporting variation data from sequencing platform, single and double channel gene expression and Copy Number Variation data in VCF, SEG, RES and GCT file formats are supported. These formats are supported by the IGV browser.
You can download data for patients or subjects already selected in an active query or from a query library or from ad-hoc list of patients ID. There is no upper limit on the number of patients or subjects to be selected, however the performance slows down as more and more patients are selected.
After selecting Patients, select the Assembly Version, Specimen Type, and Anatomical Site. These selection criteria will help you filter out patients based on the requirements. Specimen Type and Anatomical Site also have multiselect options. Currently only one version of data can be exported at a time.
You can download genomic data for either a list of genes or pathway or a gene set for a defined region in chromosome. You can also export the genomic data for a specific chromosome region and also the complete genomic data for the patient using the All Data option.
On Exadata, the code takes advantage of chromosome based partitioned data for VCF and SEG export. This enables more accurate results to be exported, including intergenic result. On non-Exadata systems, only the results that lie within any gene boundary are exported.
You can select genes from one or more of the provided three options. Using Ad-hoc List, you can select one or more genes. Using Pathway, you can select one or more pathways which in turn will get the list of genes associated with the selected pathway internally for querying. With Gene Set, you can use the user-defined collection of genes.
The genomic data to be downloaded is based on the above selected genes.
You can alternatively download genomic data using the genomic co-ordinates. You specify the chromosome region in a standard format for the Variation and CNV data to be exported. The Gene Expression - RES and Gene Expression - GCT download option would be disabled for genomic region criteria. You can specify a complete chromosome or a part of chromosome as criteria. Currently, only one chromosome region at a time is implemented for search.
The following chromosome region formats are supported.
CHR15:10000-200000: Considers region between 10000 to 200000 in chromosome 15.
CHR15:1,200,000+5000 - Considers 5000 bases upstream from 1,200,000 position in chromosome 15.
CHR15 - Considers whole of the chromosome 15.
CHR15:1000 - Considers 1000th nucleotide position of chromosome 15.
The genomic location selection is All Data option, which is only available for Schedule download and not for immediate download. With this option you can download all the data available for the specimens belonging to the selected patients or subjects falling under the selected criteria.
This panel lists out four file type options to export. You can select all four options at a time. For Genomic Region criteria, the Gene Expression – RES, and Gene Expression Dual Channel options are disabled. Once you select the option and click Submit, the data is generated and a link is provided in the bottom panel separately for each result type.
This option exports the sequencing variation data for the selected patients or subjects for either the selected genes, pathway, geneset or for a given chromosome region as selected in the previous option. VCF supports multiple specimens' data in a single file.
The metadata header gives the following information that differs based on the search criteria:
##fileformat=VCFv4.1
##fileDate: Date and time of the VCF file generated.
##source=Omics Data Bank (ODB)
##Total Number of patients included in this VCF file
##Total Number of samples included in this VCF file
7. ##INFO=<ID=NS, Number=1, Type=Integer, Description=Number of Samples With Data>
##FORMAT=<ID=GT,Number=1,Type=String,Description=Genotype>
##FORMAT=<ID=GQ, Number=1, Type=Integer, Description=Genotype Quality>
##FORMAT=<ID=GQVAF, Number=2, Type=Integer, Description=Genotype_quality_X>
##FORMAT=<ID=DP, Number=1, Type=Integer, Description=Read Depth>
##FORMAT=<ID=AD,Number=.,Type=Integer,Description=Allelic depths for the ref and alt alleles in the order listed >
##FORMAT=<ID=HQ, Number=2, Type=Integer, Description=Haplotype Quality>
##FORMAT=<ID=BQ,Number=.,Type=Integer,Description=Average base quality >
##FORMAT=<ID=MQ,Number=.,Type=Integer,Description=Average mapping quality >
##FORMAT=<ID=SS,Number=1,Type=Integer,Description=Variant status relative to non-adjacent Normal,0=wildtype,1=germline,2=somatic,3=LOH,4=post-transcriptional modification,5=unknown>
##FORMAT=<ID=SSC,Number=1,Type=Integer,Description=Somatic Score>
The following data types are imported to VCF file:
CHROM: chromosome
POS: position of the variation
ID: dbSNP ID or COSMIC ID associated with a variant
REF: reference allele
ALT: variant alleles
QUAL: not populated. Will have '.' specified in this column.
FILTER: is populated as PASS.
INFO: Not populated. Will have '.' specified in this column.
FORMAT:GT: genotypic data for each specimen.
FORMAT:GQ: genotype quality. If not value available in DB, then '.' is specified in the file.
FORMAT:GQX: mapped to GENOTYPE_QUALITY_X column.
FORMAT:DP: this stores the TotalReadCount for a specific variant.
FORMAT:AD: this stores the reference read count and Allele read count for a specific variant.
FORMAT:HQ: not populated as of now. Will have '.' specified in this column.
FORMAT:FT: this stores GENOTYPE_FILTER column value.
FORMAT:BQ: stores the RMS base quality.
FORMAT:MQ: stores the RMS mapping quality.
FORMAT:SS: stores the somatic status
FORMAT:SSC: stores the somatic status score value.
Flex field format: If any custom formats are available, they are also included in the export.
1000 Genomes VCF 4.1 conventions are followed while exporting variation data, however certain datatypes, which are non-standard, like BQ and MQ, may differ in convention for some customers since there is no standard way to represent them.
If NON_VARIANT and (or) NOCALL records exist for any given position, the zygosity is checked to determine if the format information from these tables is used.
Note:
For het-ref or half zygosity values, these other format fields are compared with the existing SEQUENCING information. This information is then used with zygosity to create the format string.The NON_VARIANT data allows for GQ, GQX, MQ, BQ and the first reference read count of AD. The NOCALL data allows for all format fields to be compared. Both NON_VARIANT and NOCALL do not support exporting flex fields. The GT value of the format string reflects the stored zygosity as follows:
| Zygosity | FORMAT string GT:GQ:GQX:BQ:MQ:AD:DP |
|---|---|
| het-ref | 1/0:99:98:38:45:20:10,10 |
| Half | 1/.:99:98:34,34:45,45:20:10,5 |
| Het-alt | 1/2:99:98:43,44:56,67:20:0,10,10 |
| Hom | 1/1:99:98:34,34:45,45:20:0,19 |
If there are no result records for any specimen, the export displays "." with no other information for the format.
There could be cases where users reload genetic information multiple times for the same specimen. This may create ambiguous values for the different fields that exist in the VCF export file. The export code deals with such ambiguous numerical values that represent the quality (that is, GQ, GQX, AD, BQ, MQ). This code now computes minimum values and ensure that the value of least confidence is reported. There could be more complex cases, for instance, if there are 2 different alleles for the same position belonging to the same specimen, or variants with same position for same specimen with different zygosity. The export code uses MIN functions on all values including all the text fields. This allows for VCF export to create a valid file that can be loaded into genome browsers.
Alternatively, you can choose not to consider data from a specific specimen or a specific file using following methods:
Using DELETE_FLG - A user may load results for a specimen more than once that can completely contradict previous results. Users can set the DELETE_FLG as 'Y' on W_EHA_RSLT_SPECIMEN and (or) W_EHA_SPEC_PATIENT or W_EHA_SPEC_SUBJECT to have previous loads excluded, and then reload the correct result files. When the user now exports the data, only the latest loaded specimen data is considered for export.
Using FILE_URI - Oracle recommends using this method since you need not reload the data again as opposed to the above method. When there are multiple files loaded with contradicting data for the same specimen, user can set some files as obsolete by changing the W_EHA_FILE_LOAD.FILE_WID column. For example, if you have loaded the same specimen data 3 times and would like to consider the latest file loaded for export, then you must first identify the latest FILE_WID from W_EHA_FILE_LOAD table. Then change the FILE_WID of two old files in W_EHA_FILE_LOAD table to the latest FILE_WID. Now, all the three records belonging to the three file loads contain same FILE_WID, which represents the latest file load and only the latest file export data is exported.
Allele depth values represented under the AD datatype are in the order of the alleles represented in the GT. Refer to the following table with examples:
| ALT | FORMAT | SAMPLE1 | |
|---|---|---|---|
| G,C,T | GT:AD | 1/2:0,4,6 | 0 represents reference_read_count
4 represents allele_read_count of 'G' 6 represents allele_read_count of 'C' |
| G,T | GT:AD | 2/2:0,4 | 0 represents reference_read_count
4 represents allele_read_count of 'T' |
| G,T | GT:AD | 1/0:10,5 | 10 represents referen ce_read_count
5 represents allele_read_count of 'G' |
The copy number variation data is exported in SEG format. Currently, CNV data from any array based system like Affymetrix Genome Wide SNP 6 array whose data is in SEG format while loading in ODB is supported. The main requirement for exporting CNV data is to have the SEG_MEAN value in the CNV table of ODB.
For exporting data that is not loaded from SEG files, for example, data from CGI CNV files or any other source of CNV data, users have to create their own loader. The loader is expected to calculate the SEG_MEAN value since this value is most important for export.
ID: specimen ID of the reported CNV segment
chrom: chromosome name
loc.start: start position of the CNV segment
loc.end: end position of the CNV segment
num.mark: for array based CNV data, this stores the number of probes details
seg.mean: this stores the segment mean value from SEG_MEAN column in CNV table.
RES is one of the gene expression formats supported by IGV browser. Currently, only microarray gene expression data is exported to this format. Following data types are imported to RES format:
Description: hugo name of a specific probe
Accession: probe ID
Intensity: intensity value of the associated probe
Call: call of the associated probe
GCT is one of the gene expression formats supported by IGV browser. Currently, only AgilentG4502A platform microarray gene expression data is exported to this format. Following data types are imported to GCT format:
Description: Gene symbol of a specific probe
Accession: probe ID
Intensity: intensity value of the associated probe
Call: call of the associated probe
Note:
The GCT file takes its gene symbol for the probe from the 2-channel composite element of ADF file. This is input into the ADF composite table in ODB. This value may not match with HUGO name in certain cases as TRC associates 2-channel records in the result table that has partial (which includes a flanking region set by the user) genomic coordinate. The coordinate overlaps between composite elements and gene segments in the reference. This may also result in some cases in more than one unique gene in the reference mapping to a gene composite element.Currently, you can export data in the following two ways:
Select option to download last loaded file(s)
Immediately, which is the default option
Schedule
Figure 5-36 Export Options - Schedule Mode
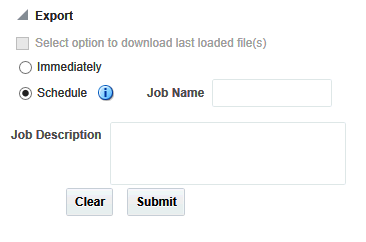
The default option (immediately) gives you the file link on the same screen and you can click to download it immediately. The link provided has a specific naming convention: <file type>_ODB_<date:MM-DD-YYYY>_<time:HH24*-MI-SS>.<file_type_extension>. For example, RES_ODB_09-14-2014_04-26.res. A short description of the file stating data type and advise on the expected count of features is displayed below the created link.
The scheduler option runs the process as a job. You can track the status of the job from the Home > Jobs tab. This option is best suited for exporting large data set like All Data, whole chromosome variants and so on. For schedule option, you must provide a job name and description. Then click Submit to start the process. For more details, see Jobs.
There is a possibility of replicate and duplicate data in the database. This could be due to loading multiple files belonging to the same specimen_number. This can happen if the same library is sequenced multiple times or the data is reanalyzed, for example, the reads were realigned using the new reference version and hence new VCF or gVCF files are created for same sample. In this scenario, you can use the option to export VCF data only from last loaded files. For example if variation data has been loaded for a specimen in Jan 2015, Mar 2015 and July 2015, then using this option you can export data from the file loaded in July 2015 and it would not consider variants from the file loaded in Jan and Mar 2015.
Note:
The Schedule Jobs option uses an asynchronous approach to store the file in DBFS. As an alternative to downloading the file using the link in the Cohort Explorer Jobs page, there are other ways to access DBFS. From a Linux OS, you can mount DBFS using dbfs_client application and then browse the directories. Windows OS does not support the FUSE interface and cannot mount DBFS directly. However, there is a dbfs_client application for Windows that can execute commands to access DBFS. The Windows version of dbfs_client lets you use the command line to execute normal directory commands. You can list the DBFS directories as well as copy data from DBFS to the local drive. The dbfs_client application is part of the standard Oracle client software.For more information about using dbfs_client, see http://docs.oracle.com/cd/E11882_01/appdev.112/e18294/adlob_client.htm#ADLOB0006.