| Oracle® Health Sciences Translational Research Center User's Guide Release 3.1.0.3 E66623-05 |
|
|
PDF · Mobi · ePub |
| Oracle® Health Sciences Translational Research Center User's Guide Release 3.1.0.3 E66623-05 |
|
|
PDF · Mobi · ePub |
This chapter contains the following topics:
ODB is a product within the Translational Research Center product family. It consists of a data model to store data as well as a set of tools that is loader scripts to load data into the model. The ODB standalone license does not include any web-based user interfaces. However, if ODB is licensed in addition to CE, several web-based User Interfaces are enabled to query against the ODB model.
ODB can be functionally thought of in two groups of tables. One set includes the reference tables which provide the metadata required to link results to specific portions of the genome. The second set represents the result tables used to capture results and link each result to an object in the reference model, and link the results back to the patient. The patient link is accomplished by linking the ODB with the Cohort Datamart, which is a part of CE 3.1.
The following sections list user interfaces available for querying ODB when licensed together with CE.
The Genomic Query tab can be utilized as a search interface into the patient genomic results in ODB. Two search modes are provided.
This tab lets you to specify one or more genes to be searched across results in ODB. You must specify Species, Assembly Version and Study that provides the output of the result data.Study is an optional criteria that you can provide in the subject context.
Assembly version restricts all the results associated against it. For Gene Search, select the Assembly version. Rather only the annotations of the selected genes are retrieved based on the Assembly version. The default Assembly version is the one that was last loaded in ODB, but you can select any one. If you select the Assembly version GRCh38, then the query retrieves gene co-ordinates for this Assembly version and a list of chromosomes present in this region.
There are currently three ways by which the genes may be specified:
Specify one or more genes explicitly by name.
Note:
When you search for a gene (hugo_name), you may also see results for a different hugo_name. This is because of synonyms or aliases present in the gene cross-reference in the ODB data model.These synonyms or aliases originate from different data sources and are loaded from HGNC HUGO data source. For example, when data loaded from HGNC Hugo has results associated with it, if you select hugo_name BNIP3 in the genomic query, you may also see results of NIP3. If you search for A1CF gene, you may see an alias ACF64.
These synonyms and alias do not appear when you search with pathway as input.
Specify gene set. If you have a set of genes that you would like to search for, you can specify the gene set and all participating genes in the gene set is searched for results.
Specify pathway to search for all genes belonging to a given pathway. Pathway information is on the reference side of the ODB model tables.
Once appropriate selections are made, click Submit to see the output information. This information includes:
Tabular list of metadata for each gene that is found and not found to have genomic results in ODB. Columns for which no data is found are labeled with No Data. The columns include:
HUGO gene name,
Ensembl gene name,
Assembly Version,
Chromosome
the number of distinct patients (or subjects) found with any data for a given gene,
the number of distinct specimen found with any data for a given gene,
the number of matching result files present in ODB
the type of result files present in ODB. For example, VCF, MAF, gene expression.
You can export the resulting table to a spreadsheet or print it. Click the Patient count to navigate to different screens like View Record, Circular Genomic Viewer, Cohort List, Cohort Timelines, Cohort Reports and Genomic Data export.
If the pathway, geneSet or genes have been selected from the ad hoc genes selection, the result details table will be paginated based on gene selection. The first page displays the result for 25 gene selection and the next page will be the result for next 25 genes and so on.
If there is no result data for the gene you are searching, then it appears in the table with No Data in the columns as specified in Figure 8-1.
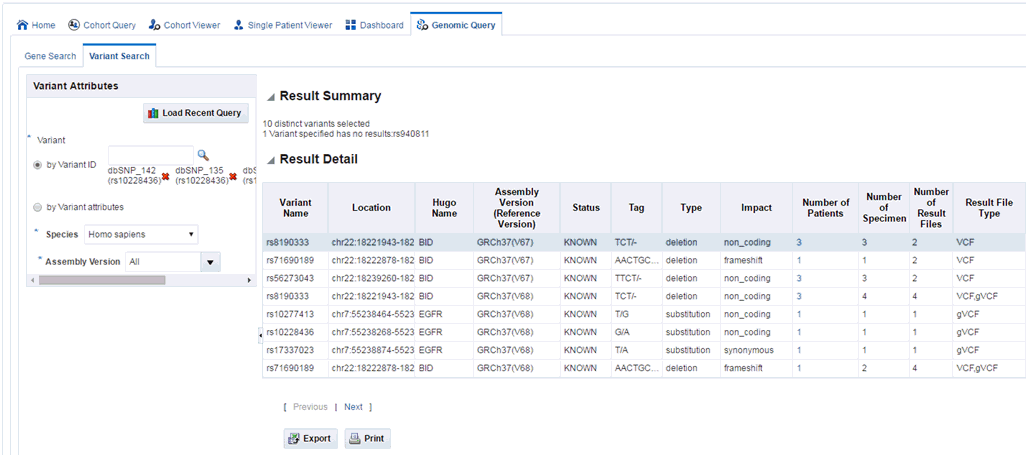
This tab lets you to specify one or more variant identifiers to be searched across results in ODB. You must specify Species and Assembly Version that displays the result data. Study can be provided as an optional parameter in the subject context.
There are currently two ways that variants may be specified:
Specify one or more variants explicitly by their variant identifier. Currently, the reference side of the schema once loaded will have Ensembl sourced variant identifier which include dbSNP and Cosmic identifiers. You can specify either one of those to try to find variants.
Specify variants by attributes like location on a gene or DNA source and additional metadata. You can provide genes in 3 ways:
Specify one or more genes explicitly by name.
Specify gene set. If you have a set of genes that you would like to search for, you can specify the gene set and all participating genes in the gene set are searched for results.
Specify pathway to search for all genes belonging to a given pathway. Pathway information is on the reference side of the ODB model tables.
You can specify Genomic Position by typing in chr#:from-to location. For example, to find variants in base pair region 1-200 on chromosome 7, you would need to type in chr7:1-200. Also, if you want to search for any variants on chromosome 7, you can just type in chr7. Additional options that can be specified include known or novel status on the variant, variant type such as insertion, deletion, substitutions, indel, complex. The strand is the directionality of the variant and it can be either plus (+) or minus (-).
Once appropriate selections are made, you can click Submit to see the output information. This information includes:
Summary of the output: this displays how many variants were searched for, how many have results found and, if applicable, how many do not have any results present in ODB for the criteria selected, what types of result files are found for the variants if any.
Tabular list of metadata for each variant that is found to have genomic results in ODB. The columns list includes the following option:
Variant Name, if the name exists,
Location of the variant,
Hugo Name
Assembly Version (Reference Version)
Status, whether the variant is known or novel,
the replace tag that tells the end user what exactly the variant is,
Type, whether this variant is an insertion, deletion and so on,
Impact, the impact of a given variant (only available for known variants),
Study, the study this variant has been found to have results for,
Number of Patients, the number of distinct patients found with any data for the particular variant,
Number of Specimen, the number of distinct specimen found with any data for the particular variant
Number of Result File, the count of type of result files present in ODB,
Result File Type, type of result files present in ODB, for example, VCF, MAF, gene expression with data for a given variant.
Note:
On gene search selections: A variant may be present in alternate genes that overlap with the gene region of the selected gene in a given version. For any variant found, these alternate gene annotations are displayed to the user along with annotations from gene of interest.You can export the resulting table to a spreadsheet or print it.