By default, the clustering process begins with all variables in a single cluster. The process is the same for each cluster and is as follows: Can we add a reliable reference to support this technique? the user can go to that source to understand the details of the technique
1. A cluster is chosen for splitting. The selected cluster should be having the smallest percentage of variation explained by its cluster component.
2. The chosen cluster is split into two clusters by finding the first two principal components, performing an orthoblique rotation (raw quartimax rotation on the eigenvectors), and assigning each variable to the rotated component with which it has the higher squared correlation.
3. Variables are iteratively reassigned to clusters to maximize the variance accounted for by the cluster components. The iterative reassignment of variables to clusters proceeds in two phases:
· The first phase is the nearest component sorting (NCS) phase. In each iteration, the cluster components are computed, and each variable is assigned to the component with which it has the highest squared correlation.
· The second phase involves a search algorithm in which each variable is tested to see if assigning it to a different cluster increases the amount of variance explained. If a variable is reassigned during the search phase, the components of the two clusters involved are recomputed before the next variable is tested.
Use the aif.feature_select_with_varclus Python API to perform variable clustering. The following are the inputs:
· X: Stage 2 converted data as a pandas data frame.
· y: Target feature as pandas series in sync with independent features X
· feature_include: Initial filter to select features from the input data.
· feature_exclude: Intialt filter to remove features from the input data.
· min_proportion: Specifies the proportion or percentage of variation that must be explained by the cluster component. The default value is 0.9.
· max_clusters: Specifies the largest number of clusters desired. Defaults to None. The default value is None.
To perform variable clustering, execute the commands as shown in the following paragraph:
|
%python aif.feature_select_with_varclus( X = Stage_2_OSIT_pdf, y = y_osit, feature_include = None, feature_exclude = None, min_proportion = 0.75, max_clusters = None ) |
The output returns the following data:
· Selected features list.
· Excluded features list.
· R square statistics.
· Cluster summary.
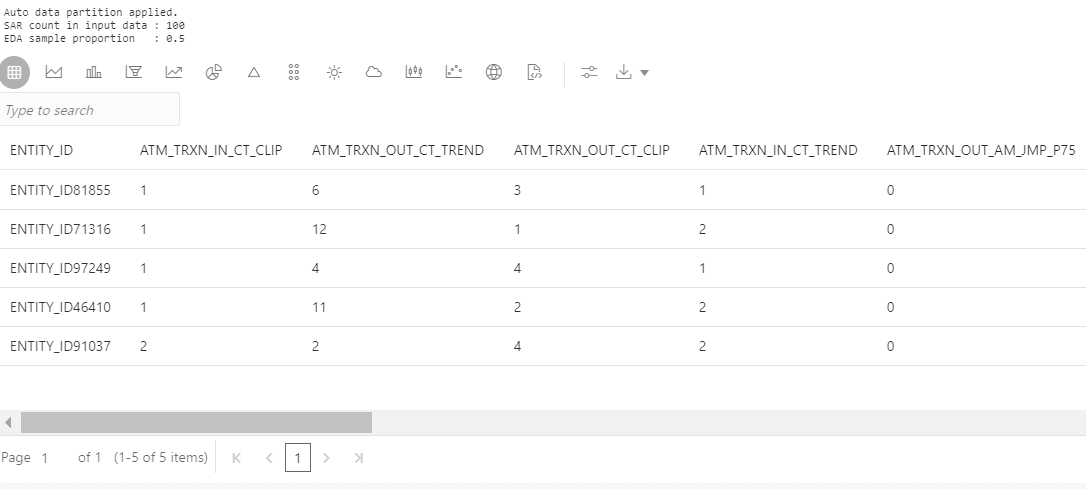
The output returns the sampled data as a pandas data frame as shown in the following paragraph:
4.
|
%python
Stage_2_OSIT_EDA_pdf = aif.eda_sampling( input_pdf = Stage_2_OSIT_pdf, eda_sample_proportion = None )
z.show( Stage_2_OSIT_EDA_pdf.head() ) |
Figure 28: Generate Data for EDA Output (Continued…)
