K-Means
K-means is a widely used clustering algorithm. It is based on proximity.
It starts with a set of cluster centroids, and each observation is assigned to the closest centroid. Then, a cluster's centroid is updated with the average of the observations assigned to it. The process continues until the centroids are no longer updated, or until a maximum number of iterations is reached.
The KMeansClustering class does not support regionalization.
This means that elements of the same clusters can be geographically disconnected.
However, you can use K-means as a benchmark for comparison purposes. It can directly
take an instance of SpatialDataFrame as input parameter for
modeling, even though the spatial information is not leveraged. It can then be
incorporated into the Spatial Pipeline.
The K-Means algorithm requires defining the number of clusters with the
n_clusters parameter. But if this is not known, the
KMeansClustering class provides two methods to estimate the number
of clusters. The user can specify any of the following methods in the
init_method parameter.
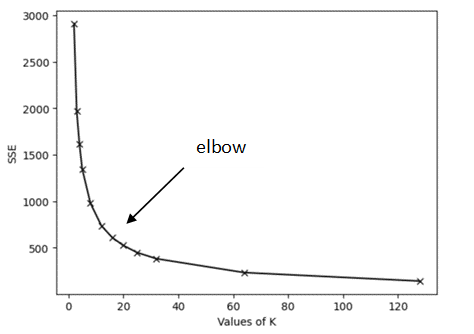
- The Elbow method: This strategy runs the K-Means algorithm for different
values of K and keeps track of the sum squared error (SSE) for each one. By plotting
the errors against the values of K, the optimal value of K is given by the graph's
"elbow" as shown in the following figure:
- The Silhouette method: This method runs the K-Means algorithm
for different values of K and measures the Silhouette score for each run. It returns
the value of K associated with the greatest score. The Silhouette score measures how
well each observation lies within its cluster. The measure is in the range
[-1, 1], and it can be interpreted as follows:- A silhouette coefficient near
1indicates that the observation is far from the neighboring clusters. - A value of
0suggests that the observation is close to or on the decision boundary between two adjacent clusters. - A negative value indicates that the observation might have been assigned to the wrong cluster.
- A silhouette coefficient near
If the n_clusters parameter is not defined, then the
algorithm uses the elbow method to estimate it. See the KMeansClustering class in Python API Reference for Oracle Spatial AI for more
information.
The following example uses the blocks_groups
SpatialDataFrame and the KMeansClustering class to
identify clusters based on specific features.
from oraclesai.weights import KNNWeightsDefinition
from oraclesai.clustering import KMeansClustering
from oraclesai.pipeline import SpatialPipeline
from sklearn.preprocessing import StandardScaler
# Define training features
X = block_groups[['MEDIAN_INCOME', 'MEAN_AGE', 'MEAN_EDUCATION_LEVEL', 'HOUSE_VALUE', 'geometry']]
# Create an instance of KMeansClustering with K=5
kmeans_model = KMeansClustering(n_clusters=5)
# Create a spatial pipeline with preprocessing and clustering steps.
kmeans_pipeline = SpatialPipeline([('scale', StandardScaler()), ('clustering', kmeans_model)])
# Train the model
kmeans_pipeline.fit(X)
# Print the labels associated with each observation
print(f"labels = {kmeans_pipeline.named_steps['clustering'].labels_[:20]}")The output consists of the labels of the first 20 observations.
labels = [3 1 3 1 3 3 1 1 3 3 2 2 1 3 2 2 1 1 1 1]