6 Configuring Data Extract
This section includes:
-
Section 6.1, "Adding the opapps User to the OCLSASCR User Group"
-
Section 6.3, "Regenerating SAS Views Created in Pre-5.0 Oracle Clinical Releases"
-
Section 6.5, "Configuring Default Installation Data Extract Settings"
-
Section 6.6, "Setting Values in Data Extract-Related Reference Codelists"
-
Section 6.7, "Creating Tablespaces for Data Extract Tables and Indexes"
-
Section 6.10, "Enabling the View Builder and Converting Views"
Oracle Clinical supports extracting Oracle and SAS views of clinical patient data. SAS is optional and must be purchased separately. The first four topics in this section apply only to SAS data extract. The remaining topics apply to both Oracle and SAS data extract.
Information on using data extract within the Oracle Clinical application is available in the Oracle Clinical Creating a Study and Oracle Clinical Conducting a Study manuals. Information on Procedures is also in Oracle Clinical Creating a Study.
6.1 Adding the opapps User to the OCLSASCR User Group
Add the opapps user to the OCLSASCR user group to give the user access to the RXC_USER directories that hold the SAS Data Extract Views. The OCLSASCR user group is created as part of the Oracle Clinical installation and has all the privileges required to use SAS. See the Oracle Clinical Installation Guide for instructions on creating the OCLSASCR group.
Individual users do not need to be members of the OCLSASCR group.
UNIX To add opapps to the OCLSASCR user group in UNIX:
-
Use the
usermodcommand, or -
Edit the /etc/group and /etc/logingroup files, if these files are not linked; if these files are linked, it is only necessary to modify the /etc/group file.
Windows You can add the user to the OCLSASCR user group in Windows by:
-
From the Start menu, navigate to Administrative Tools, then Computer Management, then Local Users and Groups, then Groups.
-
Right-click oclsascr and select Add to Group.
-
Click Add. The Select Users window opens.
-
Enter the the username and click OK.
6.2 Authenticating the SAS Connection
When you run a SAS Data Extract job, you actually run two jobs—in the first job Oracle Clinical creates a SAS file, and in the second job SAS creates a SAS view from the file created in first job.
Note:
For command line execution of the SAS file, the user must log in as opapps.In the first job, Oracle Clinical generates a connect string based on the specified authentication method to be used when the SAS file is executed.
There are two authentication methods and at any point in time, all DX jobs and SAS files must use the same option. You must enter the SAS_CONNECTION value in the OCL_STATE local reference code list: either ORACLE_WALLET or SAS_ENCRYPTION. The default value is ORACLE_WALLET.
Note:
Plan to continue to use a single option consistently. Each time you change from one to the other you must regenerate all files generated using the other option so users can view them. Also, at any point in time, all SAS jobs must use the same option.6.2.1 Using Oracle Wallet for SAS Authentication
This is the default option. The connect string generated for Oracle Wallet is:
connect to oracle(user='oc_end_user 'password=''path='dbname');
Although the individual user ID is specified in the file, the connection is actually made with this account's grant to the ocpsub proxy account, which is stored in the Wallet.
-
If your SAS server is the same as the database server, and you set ORACLE_WALLET as the authentication method, the SAS connection is set up automatically—opapps retrieves the OCPSUB database password from the opapps wallet
-
If your SAS server is on a different machine from the database and you want to use ORACLE_WALLET, you must first set up the Oracle Wallet on the SAS server, as described in the Oracle Clinical Installation Guide.
6.3 Regenerating SAS Views Created in Pre-5.0 Oracle Clinical Releases
SAS views generated in earlier releases are not usable in Oracle Clinical Release 5.0 and later, because 5.0 and later require a different connection string to be embedded in the SAS files. You must re-run all jobs to create new *.sas files. Use the utility gen_views for this purpose; see "Generating Data Extract Views".
6.4 Creating SAS Output File Directories
You must create a root directory in which to store output files for the jobs SAS Datasets and Proc Print, and enter the path as the value for SAS_OUTPUT_ROOT in the OCL_STATE local reference codelist.
You must also decide whether or not you want to have a subdirectory for each user who submits these jobs, and enter either Y or N as the value for SAS_USERDIRS in the OCL_STATE local reference codelist. The system enforces the choice you make.
If you have very few users who submit SAS Datasets and Proc Print jobs, and if they all have access to the same studies, you may choose not to have user-specific subdirectories. Otherwise, having user-specific directories is the more secure choice because users can see only output files of jobs they have submitted.
If you choose to have user-specific subdirectories, you must create the subdirectories.
6.5 Configuring Default Installation Data Extract Settings
In the DX Installation Configuration window, you determine the default settings for new studies. To launch the Data Extract Installation Configuration window, navigate to Admin and select DX Installation Configuration.
The DX_CONFIG installation reference codelist includes exactly the same settings. When you change a setting here, the change is reflected in the reference codelist. When you change a setting there, the change is reflected here.
Figure 6-1 Data Extract Installation Configuration Window
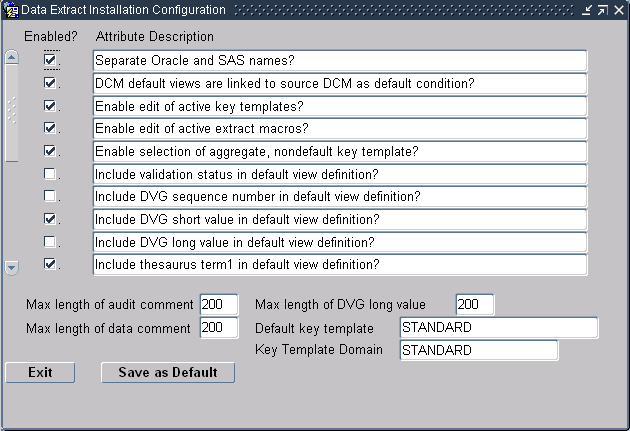
Description of ''Figure 6-1 Data Extract Installation Configuration Window''
You can enable or disable the attributes described in the upper part of the window by selecting or clearing the appropriate box. The lower part of the window enables you to reduce the size of comments and the DVG long value, and to choose the default Key Template and the default Key Template domain.
The settings in the Data Extract Installation Configuration window are:
Separate Oracle and SAS Names?
When enabled, you can specify different names for Oracle and SAS view columns. Oracle views take the long name; SAS takes the short name. The default is deselected.
In earlier versions of SAS (such as version 6.12), the maximum length for variables (columns or views) was 8 characters, while Oracle names could be as long as 30 characters. If you wanted to keep the same names for the Oracle and SAS variables (for consistency or some other business need), you had to choose a name short enough to fit in the SAS variable length. If you wanted to have a longer Oracle name, the names had to be different. Data Extract creates the Oracle and SAS views based on the decision you make in this field.
DCM Default Views Are Linked to Source DCM as Default Condition?
This setting controls whether a view definition is linked to its source DCM if the view definition's link mode is DEFAULT. If this setting is enabled, a view definition with DEFAULT link mode will be linked to its DCM, meaning that changes to the DCM will propagate to the view definition as well. If this setting is not enabled, these view definitions are not linked, so they will not change when the source DCM changes.
Enable Edit of Active Key Templates?
Enable Edit of Active Extract Macros?
Enable Edit of Active View Templates?
Each of these settings enables you to choose whether users can modify one type of active component in a view definition. Your organization may want to freeze definitions like Key Templates, extract macros, and View Templates that are used across many view definitions in the global library.
Enable Selection of Aggregate, Nondefault Key Template?
Choose this setting to be able to choose alternative Key Templates for different view definitions within a study.
Include Validation Status in Default View Definition?
Include DVG Sequence Number in Default View Definition?
Include DVG Short Value in Default View Definition?
Include DVG Long Value in Default View Definition?
Include Thesaurus Term1 in Default View Definition?
Include Thesaurus Term2 in Default View Definition?
Include Thesaurus Term3 in Default View Definition?
Include Full Value Text in Default View Definition?
These settings all control attributes that you might want to include in the default view definition. All are part of what you can add through the Extended Attributes button when defining a simple question in the Global Library, or through the Template Attributes button when building a template in the Maintain View Templates window.
-
Validation Status is an attribute of the RESPONSES table. By choosing to display another attribute, you can tell how clean your data is. The default is deselected.
-
The DVG Sequence Number indicates the order the discrete values appear in the list of values for data entry. The DVG Short Value is the data as entered. The DVG Long Value is a longer form than the short value of the data as entered. You need at least one, but you may pick all, of the following: the DVG Long Value, the DVG Sequence Number, or data values for the DVG questions.
-
The Thesaurus Term configuration preferences involve the same kinds of choices as for the DVG, except that data can come from several different tables. You must still choose at least one term, and you may choose all three. The default is selected.
-
Oracle Clinical stores valid responses in the Value Text field, and invalid ones in the Exception Value Text field. When the response is valid, the Full Value Text field contains the Value Text; when it is invalid, Full Value Text contains the Exception Value Text.
Enable Update of SAS and Oracle Column Names?
When you bring a question from the Global Library into a View Template, the SAS column names and Oracle column names in the View Template default to the names defined in the Global Library. If this option is selected, you can change the names at the View Template level; if not selected, you cannot modify the names from their Global Library-derived defaults. The default setting is deselected.
Enable View Builder as Default in New Studies?
The View Builder enables you to automatically generate views of the data and metadata included in a single Data Collection Module (DCM). If this setting is enabled, the VB Enabled? setting in the Clinical Study States window is selected by default for new studies.
Use DCM Question-Specific DVG Subset for DVG Attributes?
This setting determines whether the Discrete Value Group (DVG) attributes that are included in the view come from the DVG subset that has been assigned to the DCM question or from the base DVG subset. Selecting this box makes the views include the DCM question DVG's subset information; clearing the box makes the views include the base subset information. For information on DVGs and DVG subsets, see the chapter on questions in Oracle Clinical Creating a Study.
Use DCM SAS Label as Seed for Attributes in Default View Definition?
This setting determines which SAS labels the system uses for all the attribute columns of a default view definition. By default, this setting is not enabled, so the SAS labels of the attribute columns are created using the SAS label of the question attributes in the Global Library. However, when you enable this setting, the system creates the SAS labels of the attribute columns of the View Template within the context of a view definition by using the corresponding DCM question's SAS label as the seed when the View Template Question is mapped.
The default Audit Comment length is 200 characters. You can reduce this value if you typically use no more than a few characters for this comment.
The Oracle Clinical default Comment length is 200 characters. You can reduce this value if you typically use no more than a few characters for this comment.
The default DVG Long Value length is 200 characters. You can reduce this value if you typically use no more than a few characters for this comment.
You can also create the DVG Long Value column with a maximum width equal to the DVG values specified for a given question. This behavior is enabled when the maximum length of the DVG Long Value is set to zero.
The default Key Template for custom and default view definitions. You can choose a new default Key Template from the list of values.
Data extract users can choose a non-default Key Template for their view definition only if the Enable Selection of Aggregate, Nondefault Key Template? box is selected.
Note that study-specific Key Templates achieve the same goal. You can supply a study-specific Key Template in the Clinical Study States window (from the Conduct menu, select Security, then select Clinical Study States).
The Key Template Domain indicates the Global Library domain in which the default Key Template is stored. You cannot assign or change the domain of the default Key Template in this window.
Fast views are created with a different structure from other data extract views. While functionally equivalent, the fast view structure provides better performance when querying the views, especially for queries against response values. If set to Y, the system builds fast views when possible but builds regular views if the view structure is incompatible with the fast view approach. This is the case with cross-DCM views and with views based on key templates that aggregate across key columns such as patient, visit or received DCM. The system handles cross-DCM views automatically, but for aggregate views you must enter the text "AGGREGATE" in the Status Comment field of the key template. The system then successfully builds a regular view structure.
If you are encountering performance issues--queries are running for several minutes--when querying large data extract views, you should consider using fast views.
6.6 Setting Values in Data Extract-Related Reference Codelists
Check the settings of the following reference codelists, described in Chapter 7, "Reference Codelists":
6.7 Creating Tablespaces for Data Extract Tables and Indexes
This section includes:
6.7.1 Creating Tablespaces
You must create tablespaces to contain the tables and indexes required for data extract. Oracle recommends creating a separate tablespace for tables and for indexes, and for creating each tablespace:
-
as locally managed
-
with autoallocate on
-
with automatic segment space management
-
with a block size of 16 kb
Use the following command:
CREATE TABLESPACE dxtables DATAFILE '/u02/oracle/data/dxtables01.dbf' SIZE 500M EXTENT MANAGEMENT LOCAL AUTOALLOCATE SEGMENT SPACE MANAGEMENT AUTO;
For more information, see the Oracle Database Administrator's Guide.
6.7.2 Entering Tablespace Names in Reference Codelists
To enable users to create tables using the Data Extract View Builder, you must specify to which tablespace and tablespace index you want to add tables for each Study Access Account. You specify these tablespaces and index tablespaces in two installation reference codelists that store data extract tablespace information:
Note:
You must also remove invalid values from the reference codelists to prevent users from selecting them for study access accounts. See "DX_INDEX_TABLESPACE Installation Codelist" and "DX_VIEW_TABLESPACE Installation Codelist" for information.6.7.3 Creating Data Extract Access Accounts Using Local Tablespaces
You must create a Data Extract Access Account (under Conduct, navigate to Data Extract, then Study Access Accounts) and specify a tablespace for the account's tables and for its indexes. When the tablespace is defined as locally managed, the View Builder creates tables and indexes for that account as follows:
-
The extract tables are created in a single step rather than the previous approach that created a temporary table and then, after determining the size of the table, a second permanent table. This feature capitalizes on the ability to use locally managed tablespaces with the AUTOALLOCATE feature that automatically sizes the table.
-
The tables are created using the Oracle database table compression feature. Since data extract tables tend to have many repeated keys and values, this should result in significantly less space usage and more efficient data access.
-
The indexes are created with leading key compression that results in significantly more compact and efficient indexes.
-
Index statistics are now computed as the indexes are created, which results in faster statistics calculation.
-
Repeated account maintenance of the ROLLSNAP account will take advantage of the more efficient space allocation method used in Locally Managed Tablespaces when it drops and recreates tables.
-
Tables are created with the NOLOGGING attribute. This reduces the table creation time significantly by avoiding writing to the redo logs. The price of this option is that recovery from database failure using redo logs will not recreate extract tables that have not been otherwise backed up and restored. Since the tables can be recreated from the extract views at any time by rerunning account maintenance in FULL, this trade-off is usually acceptable. However, if you do not want to use the NOLOGGING behavior, you can override it by creating the locally managed tablespace with the FORCE LOGGING attribute.
6.8 Customizing Data Extract Views
Oracle Clinical ships with two scripts in the INSTALL directory that you can use to customize data extract views.
-
rxcptdxvb.sql populates the data extract tables EXTRACT_KEYS and EXTRACT_MACROS and creates the standard key template.
-
pop_vb_static_views.sql creates the standard view templates, which include views for responses and RDCMs.
For example, follow these instructions to make the investigator's last name available to an extract macro:
-
Back up the scripts in the directory $RXC_INSTALL that are used to customize data extract views:
-
pop_vb_static_views.sql
-
rxcptdxvb.sql
-
-
Bring up pop_vb_static_views.sql in a text editor.
-
Make the changes marked in bold so that the updated code is as shown below:
… REM REM ORACLE TEXT FOR NEW STYLE RDCMS_VIEW REM DECLARE LTEXT long; BEGIN /* SPR24128 JRees 9/24/98 Adding hints to security checks */ LTEXT := 'create view \0.rdcms_view as select /*+ ORDERED USE_MERGE(css) INDEX(rdcm RECEIVED_DCM_DCM_CS_NFK_IDX\4) INDEX(css CLINICAL_STUDY_STATE_IDX) <- Remove '*/' from the end of this line. /* to add additional tables, add index hint for joining */ /* for example, for RDCIs add: */ /* (note that \4 adds the T for test mode) */ /* INDEX(RDCI RECEIVED_DCI_PK_IDX\4) */ /* for example, for OCL_Investigators, add: */ INDEX(INV OCL_INVESTIGATOR\4_PK_IDX) */ <- Remove '/*' from the start of this line. rdcm.received_dcm_id, rdcm.dcm_id, rdcm.dcm_subset_sn, rdcm.dcm_layout_sn, rdcm.actual_event_id, rdcm.dci_id, rdcm.received_dci_id, rdcm.received_dcm_entry_ts, rdcm.end_ts, rdcm.entered_by, rdcm.dcm_date, rdcm.dcm_time, rdcm.received_dcm_status_code, rdcm.qualifying_value, rdcm.accessible_ts, rdcm.log_in_ts, rdcm.last_data_change_ts, rdcm.data_lock_flag, rdcm.sn, rdcm.document_number, rdcm.modification_ts, rdcm.modified_by, rdcm.subevent_number, rdcm.investigator_id, rdcm.investigator, inv.last_name, <- Add this line. Remember the comma. rdcm.clin_plan_eve_id, rdcm.clin_plan_eve_name, rdcm.visit_number, rdcm.site_id, rdcm.site, rdcm.lab_id, rdcm.lab, rdcm.lab_range_subset_num, rdcm.LAB_ASSIGNMENT_TYPE_CODE, rdcm.patient_position_id, rdcm.patient, rdcm.clinical_study_id /* to add additional columns, add here, */ /* for example, for Received_DCIs, add: */ /* , RDCI.FIRST_BOOK_PAGE */ /* for example, for OCL_Investigators, add: */ /* , INV.COUNTRY */ \5 /* to customize, extend from list. */ /* for example, to add RDCIs add: */ /* note: the \4 adds the T for test account */ /* , RECEIVED_DCIS\4 RDCI */ /* for example, to add OCL_Investigators add: */ , OCL_INVESTIGATORS\4 INV <- Remove '/*' and '*/' from this line. WHERE \1 /* use \6 to access as_of_ts for account-specific */ /* time restriction, */ /* for example, to join RDCIS, add: */ /* AND RDCM.RECEIVED_DCI_ID = RDCI.RECEIVED_DCI_ID */ /* AND RDCI.END_TS > \6 */ /* AND RDCI.RECEIVED_DCI_ENTRY_TS <= \6 */ /* for example, to join OCL_Investigators, add: */ AND RDCM.INVESTIGATOR_ID = INV.INVESTIGATOR_ID <- Remove '/*' and '*/' from this line. and (exists /* account is super-user */ (select /*+ index(oa ORACLE_ACCOUNT_PK_IDX) */ … -
Save the changes made to the pop_vb_static_views.sql file.
-
To add the column as an Extract Key, bring up rxcptdxvb.sql in a text editor.
-
Make the changes marked in bold:
delete from extract_keys where OC_INTERNAL_NAME in ( '/STUDY', 'DCMS.SUBSET_NAME', 'DCMS.SUBSET_NAME', 'DCMS.DCM_SUBSET_SN', 'RDCM.DOCUMENT_NUMBER', 'RDCM.SITE', 'RDCM.INVESTIGATOR', 'RDCM.LAST_NAME', <- Add this line. 'RDCM.PATIENT', 'RDCM.ACCESSIBLE_TS', 'RDCM.LOG_IN_TS', 'RDCM.LAST_DATA_CHANGE_TS', 'RDCM.DATA_LOCK_FLAG', 'RDCM.CLIN_PLAN_EVE_NAME', 'RDCM.DCM_DATE', 'RDCM.DCM_TIME', 'R.REPEAT_SN', …Note:
The new key is added as RDCM.LAST_NAME and not INV.LAST_NAME. In this context RDCM is the alias for the RDCMS_VIEW view to which we added the new column by modifying pop_vb_static_view.sql.The rxcptdxvb.sql file already contains the required modifications for adding FIRST_BOOK_PAGE from RECEIVED_DCIS and COUNTRY from OCL_INVESTIGATORS. No further modification of this file is therefore required for these columns if you have added them to pop_vb_static_view.sql.
-
Find the
insert into extract_keysstatement for RDCM.DCM_DATE and duplicate it to create an insert statement for the RDCI.INVESTIGATOR. -
Save the changes made to the rxcptdxvb.sql file.
-
To populate the data extract tables with the customized definitions, log in to SQL*Plus as RXC and run both scripts:
SQL> start pop_vb_static_views.sql SQL> start rxcptdxvb.sql
Note:
Note: If when running rxcptdxvb.sql you getORA-0001: Unique constraint (RXC.EXTRACT_KEYS_ORA_UK_ID) violatedthen perform the following as RXC to clean up:SQL> delete extract_keys; SQL> commit;
Re-execute rxcptdxvb.sql and continue with the next step.
-
To confirm that the INVNAME extract key and macro are now available for use, Log into Oracle Clinical and navigate to Glib, then Data Extract View Builder, then Extract Macros and press F8 to query. INVNAME should now be included.
-
To add the extract key and extract macro for the new column to a Key Template, . Query for the template to which
-
Navigate to Glib, then Data Extract View Builder, then Key Templates.
-
Query for the template to which you want to add the macro.
-
Click on Key Columns.
-
In an empty field, press F9 to see the list of values.
-
Select INVNAME from the list.
-
Save.
-
The investigator's last name will appear as column INVNAME in all data extract views created with this Key Template.
6.9 Generating Data Extract Views
The gen_views utility performs the same operations as the Maintain Data Extract Views batch job within Oracle Clinical (under Conduct select Data Extract and then Data Extract Views) for all accounts in FULL maintenance mode.
Use the gen_views utility to regenerate views for all accounts in one or all studies. For example, if a change is made to a key template, all views based on that template in that study must be regenerated for all the study access accounts.
The general sequence for this task is:
-
run opa_setup, which defines the RXC_TOOLS directory
-
change to the RXC_TOOLS directory
-
run gen_views.
Variables include:
-
study enter either the name of a study or ALL for all studies
-
sas_queue enter the name of the queue where SAS jobs execute or NULL (on Windows only)
-
view_creation_mode valid values are: DATA_ONLY, COMBINED_VIEW, or SEPARATE_VIEW.
Operating system-specific instructions follow:
6.9.1 Running gen_views on UNIX Platforms
To run gen_views on a UNIX platform:
-
Log on to the server in your user account.
-
Set environment variables for the database name and code environment; see "Setting Environment Variables on the Command Line."
-
Change directories to $RXC_TOOLS.
-
Set the output directory:
-
C Shell command:
setenvRXC_LOG usr_log_dir -
Bourne Shell command:
RXC_LOG=usr_log_direxport= code_env
-
-
Run the script:
gen_viewsstudyUNIXview_creation_modeFor example:
gen_views ALL UNIX DATA_ONLY
The script prompts for:
-
Database name
-
Username for user who can submit the DX job
-
Password for the user
-
6.9.2 Running gen_views on Windows
To run gen_views on Windows:
-
Log on to the server using your local account.
-
In an MS-DOS window, set the server environment:
set p1=db_name
set p2=code_env
opa_setup
where db_name is a database instance name and code_env is a code environment designation.
-
Change directories to %RXC_TOOLS%
-
Set the output directory:
set rxc_log=user_log_folder
-
Run the command file.
gen_viewsstudyNULLview_creation_modeFor example:
gen_views ALL NULL DATA_ONLY
The script prompts for:
-
Database name
-
Username for user who can submit the DX job
-
Password for the user
-
6.10 Enabling the View Builder and Converting Views
The View Builder was an enhancement in Oracle Clinical Release 3.1. If you are still using pre-View Builder views, you can convert them to View Builder-style views by running two scripts, vb_pop_view and enable_vb, for each study.
6.10.1 Converting Views
Running vb_pop_view converts a study's existing old-style data extract views to the new view builder style. For example:
cd $RXC_INSTALL sqlplus rxc/notrxc start vb_pop_view
The script prompts you for the name of the study.
Note:
You can run vb_pop_view only once per study.6.10.2 Enabling the View Builder in a Study
To make view builder-style the default for future views in a study, use the enable_vb.sql script. For example:
cd $RXC_INSTALL sqlplus rxc/notrxc start enable_vb.sql
When you execute this script it performs the following actions:
-
prompts you for the name of the study.
-
updates the CLINICAL_STUDY_STATES table, setting VB_ENABLED to "Y".
-
sets the default key template in CLINICAL_STUDY_STATES to STANDARD.
After running enable_vb, commit the changes to the database, then rerun view creation.
You can also enable view builder interactively for individual studies within the Oracle Clinical interface. However, using the command line may be more convenient.
6.11 Controlling Access to Data Extract Views
Access to individual views at the access account level can be controlled by granting access via roles. You can create your own company-specific roles so that appropriate choices appear in the list of values in the View Definition window.
Do the following:
-
Create as many database roles as you need; see "Creating Custom Database Roles".
-
Add these roles to the DX_ROLES Installation Codelist. All the roles in that reference codelist appear in the list of values in the View Definition window.
-
Grant the database roles to users who need them; see "Granting Additional Database Roles to User Accounts".