4.2.1 About Building Oracle Data Mining Models using Oracle R Enterprise
Oracle Data Mining can mine tables, views, star schemas, transactional data, and unstructured data. The OREdm functions provide R interfaces that use arguments that conform to typical R usage for corresponding predictive analytics and data mining functions.
This section has the following topics:
4.2.1.1 Oracle Data Mining Models Supported by Oracle R Enterprise
The functions in the OREdm package provide access to the in-database data mining functionality of Oracle Database. You use these functions to build data mining models in the database.
The following table lists the Oracle R Enterprise functions that build Oracle Data Mining models and the corresponding Oracle Data Mining algorithms and functions.
Table 4-2 Oracle R Enterprise Data Mining Model Functions
| Oracle R Enterprise Function | Oracle Data Mining Algorithm | Oracle Data Mining Function |
|---|---|---|
|
|
Minimum Description Length |
Attribute Importance for Classification or Regression |
|
|
Apriori |
Association Rules |
|
|
Decision Tree |
Classification |
|
(12.2 feature) |
Expectation Maximization |
Clustering |
|
(12.2 feature) |
Explicit Semantic Analysis |
Feature Extraction |
|
|
Generalized Linear Models |
Classification and Regression |
|
|
k-Means |
Clustering |
|
|
Naive Bayes |
Classification |
|
|
Non-Negative Matrix Factorization |
Feature Extraction |
|
|
Orthogonal Partitioning Cluster (O-Cluster) |
Clustering |
|
(12.2 feature) |
Extensible R Algorithm |
Association Rules, Attribute Importance, Classification, Clustering, Feature Extraction, and Regression |
|
(12.2 feature) |
Singular Value Decomposition |
Feature Extraction |
|
|
Support Vector Machines |
Classification and Regression |
4.2.1.2 About Oracle Data Mining Models Built by Oracle R Enterprise Functions
In each OREdm R model object, the slot name (or fit.name) is the name of the underlying Oracle Data Mining model generated by the OREdm function. While the R model exists, the Oracle Data Mining model name can be used to access the Oracle Data Mining model through other interfaces, including:
-
Oracle Data Miner
-
Any SQL interface, such as SQL*Plus or SQL Developer
In particular, the models can be used with the Oracle Data Mining SQL prediction functions.
With Oracle Data Miner you can do the following:
-
Get a list of available models
-
Use model viewers to inspect model details
-
Score appropriately transformed data
Note:
Any transformations performed in the R space are not carried over into Oracle Data Miner or SQL scoring.
Users can also get a list of models using SQL for inspecting model details or for scoring appropriately transformed data.
By default, models built using OREdm functions are transient objects; they do not persist past the R session in which they were built unless they are explicitly saved in an Oracle R Enterprise datastore. Oracle Data Mining models built using Data Miner or SQL, on the other hand, exist until they are explicitly dropped.
Model objects can be saved or persisted. Saving a model object generated by an OREdm function allows it to exist across R sessions and keeps the corresponding Oracle Data Mining object in place. While the OREdm model exists, you can export and import it; then you can use it apart from the Oracle R Enterprise R object existence.
You can use the MODEL_NAME parameter in odm.settings to explicitly name an Oracle Data Mining object created in the database. The named Oracle Data Mining model object persists in the database just like those created using Oracle Data Miner or SQL.
Related Topics
4.2.1.3 Partitioning and Text Mining
Beginning with Oracle Database 12c, Release 2 (12.2), functions in the Oracle Data Mining package have an argument that specifies settings for an Oracle Data Mining model and some have an argument for setting text mining parameters.
With the odm.setting argument to an OREdm function, you can specify a list of Oracle Data Mining parameter settings. Each list element's name and value refer to the parameter setting name and value, respectively. The setting value must be numeric or string. Refer to Oracle Data Mining User’s Guide for each algorithm's valid settings.
The settings function returns a data.frame that lists each Oracle Data Mining parameter setting name and value pair used to build the model.
Partitioned Oracle Data Mining Models
A partitioned model is an ensemble model that consists of multiple sub-models. To create a partitioned Oracle Data Mining model, use the odm.setting argument with ODMS_PARTITION_COLUMNS as the name and with the names of the columns by which to partition the input data as the value. The OREdm function returns a model with a sub-model for each partition. The partitions are based on the unique values found in the columns.
Partitioned models can automate scoring by allowing you to reference the top-level model only, which causes the proper sub-model to be chosen based on the values of the partitioned column or columns for each row of data to be scored.
The partitions function returns an ore.frame that lists each partition of the specified model object and the associated partition column values of the model. Partition names are system-determined. The function returns NULL for a non-partitioned model.
Text Mining Attribute Settings
Some OREdm functions have a ctx.settings argument that specifies text mining attribute settings with which you can specify Oracle Text attribute-specific settings. With the odm.settings argument, you can specify the Oracle text policy, the minimal number of documents in which each token occurs, and the maximum number of distinct features for text mining. With the ctx.settings argument, you specify the columns that should be treated as text and the type of text transformation to apply.
The ctx.settings argument applies to the following functions:
-
ore.odmESA, Explicit Semantic Analysis -
ore.odmGLM, Generalized Linear Models -
ore.odmKMeans, k-Means -
ore.odmNMF, Non-Negative Matrix Factorization -
ore.odmSVD, Singular Value Decomposition -
ore.odmSVM, Support Vector Machines
Note:
To create an Oracle Text policy, the user must have theCTXSYS.CTX_DDL privilege.See Also:
Creating a Model that Includes Text Mining in Oracle Data Mining User’s Guide for valid text attribute values.Example 4-8 Example of Text Mining with ore.odmKMeans
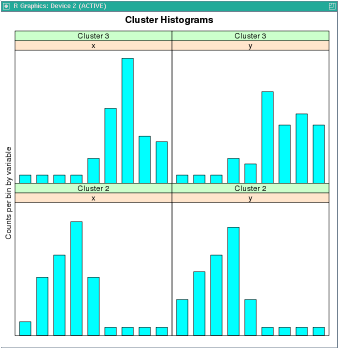
This example uses the odm.settings and ctx.settings arguments. The figure following the example shows the output of the histogram(km.mod1) function.
x <- rbind(matrix(rnorm(100, sd = 0.3), ncol = 2),
matrix(rnorm(100, mean = 1, sd = 0.3), ncol = 2))
colnames(x) <- c("x", "y")
X <- ore.push (data.frame(x))
km.mod1 <- NULL
km.mod1 <- ore.odmKMeans(~., X, num.centers = 2)
km.mod1
summary(km.mod1)
rules(km.mod1)
clusterhists(km.mod1)
histogram(km.mod1)
km.res1 <- predict(km.mod1,X,type="class",supplemental.cols=c("x","y"))
head(km.res1,3)
km.res1.local <- ore.pull(km.res1)
plot(data.frame(x = km.res1.local$x,
y = km.res1.local$y),
col = km.res1.local$CLUSTER_ID)
points(km.mod1$centers2, col = rownames(km.mod1$centers2), pch = 8, cex=2)
head(predict(km.mod1,X))
head(predict(km.mod1,X,type=c("class","raw"),supplemental.cols=c("x","y")),3)
head(predict(km.mod1,X,type="raw",supplemental.cols=c("x","y")),3)
# Text mining with ore.odmKMeans
title <- c('Aids in Africa: Planning for a long war',
'Mars rover maneuvers for rim shot',
'Mars express confirms presence of water at Mars south pole',
'NASA announces major Mars rover finding',
'Drug access, Asia threat in focus at AIDS summit',
'NASA Mars Odyssey THEMIS image: typical crater',
'Road blocks for Aids')
response <- c('Aids', 'Mars', 'Mars', 'Mars', 'Aids', 'Mars', 'Aids')
# TEXT contents in character column
KM_TEXT <- ore.push(data.frame(CUST_ID = seq(length(title)),
RESPONSE = response, TITLE = title))
# Create text policy (CTXSYS.CTX_DDL privilege is required)
ore.exec("Begin ctx_ddl.create_policy('ESA_TXTPOL'); End;")
# Specify POLICY_NAME, MIN_DOCUMENTS, MAX_FEATURES and
# text column attribute specification
km.mod <- ore.odmKMeans( ~ TITLE, data = KM_TEXT, num.centers = 2L,
odm.settings = list(ODMS_TEXT_POLICY_NAME = "ESA_TXTPOL",
ODMS_TEXT_MIN_DOCUMENTS = 1,
ODMS_TEXT_MAX_FEATURES = 3,
kmns_distance = "dbms_data_mining.kmns_cosine",
kmns_details = "kmns_details_all"),
ctx.settings = list(TITLE = "TEXT(TOKEN_TYPE:STEM)"))
summary(km.mod)
settings(km.mod)
print(predict(km.mod, KM_TEXT, supplemental.cols = "RESPONSE"), digits = 3L)
ore.exec("Begin ctx_ddl.drop_policy('ESA_TXTPOL'); End;")
Listing for This Example
R> x <- rbind(matrix(rnorm(100, sd = 0.3), ncol = 2),
+ matrix(rnorm(100, mean = 1, sd = 0.3), ncol = 2))
R> colnames(x) <- c("x", "y")
R>
R> X <- ore.push (data.frame(x))
R> km.mod1 <- NULL
R> km.mod1 <- ore.odmKMeans(~., X, num.centers = 2)
R> km.mod1
Call:
ore.odmKMeans(formula = ~., data = X, num.centers = 2)
Settings:
value
clus.num.clusters 2
block.growth 2
conv.tolerance 0.01
details details.all
distance euclidean
iterations 3
min.pct.attr.support 0.1
num.bins 10
random.seed 0
split.criterion variance
odms.missing.value.treatment odms.missing.value.auto
odms.sampling odms.sampling.disable
prep.auto ON
R> summary(km.mod1)
Call:
ore.odmKMeans(formula = ~., data = X, num.centers = 2)
Settings:
value
clus.num.clusters 2
block.growth 2
conv.tolerance 0.01
details details.all
distance euclidean
iterations 3
min.pct.attr.support 0.1
num.bins 10
random.seed 0
split.criterion variance
odms.missing.value.treatment odms.missing.value.auto
odms.sampling odms.sampling.disable
prep.auto ON
Centers:
x y
2 -0.07638266 0.04449368
3 0.98493306 1.00864399
R> rules(km.mod1)
cluster.id rhs.support rhs.conf lhr.support lhs.conf lhs.var lhs.var.support lhs.var.conf predicate
1 1 100 1.0 92 0.86 x 86 0.2222222 x <= 1.2209
2 1 100 1.0 92 0.86 x 86 0.2222222 x >= -.6188
3 1 100 1.0 86 0.86 y 86 0.4444444 y <= 1.1653
4 1 100 1.0 86 0.86 y 86 0.4444444 y > -.3053
5 2 50 0.5 48 0.96 x 48 0.0870793 x <= .4324
6 2 50 0.5 48 0.96 x 48 0.0870793 x >= -.6188
7 2 50 0.5 48 0.96 y 48 0.0893300 y <= .5771
8 2 50 0.5 48 0.96 y 48 0.0893300 y > -.5995
9 3 50 0.5 49 0.98 x 49 0.0852841 x <= 1.7465
10 3 50 0.5 49 0.98 x 49 0.0852841 x > .4324
11 3 50 0.5 50 0.98 y 49 0.0838225 y <= 1.7536
12 3 50 0.5 50 0.98 y 49 0.0838225 y > .2829
R> clusterhists(km.mod1)
cluster.id variable bin.id lower.bound upper.bound label count
1 1 x 1 -0.61884662 -0.35602715 -.6188466:-.3560272 6
2 1 x 2 -0.35602715 -0.09320769 -.3560272:-.0932077 17
3 1 x 3 -0.09320769 0.16961178 -.0932077:.1696118 15
4 1 x 4 0.16961178 0.43243125 .1696118:.4324312 11
5 1 x 5 0.43243125 0.69525071 .4324312:.6952507 8
6 1 x 6 0.69525071 0.95807018 .6952507:.9580702 17
7 1 x 7 0.95807018 1.22088965 .9580702:1.2208896 18
8 1 x 8 1.22088965 1.48370911 1.2208896:1.4837091 4
9 1 x 9 1.48370911 1.74652858 1.4837091:1.7465286 4
10 1 y 1 -0.89359597 -0.59946141 -.893596:-.5994614 2
11 1 y 2 -0.59946141 -0.30532685 -.5994614:-.3053269 4
12 1 y 3 -0.30532685 -0.01119230 -.3053269:-.0111923 11
13 1 y 4 -0.01119230 0.28294226 -.0111923:.2829423 24
14 1 y 5 0.28294226 0.57707682 .2829423:.5770768 13
15 1 y 6 0.57707682 0.87121138 .5770768:.8712114 12
16 1 y 7 0.87121138 1.16534593 .8712114:1.1653459 26
17 1 y 8 1.16534593 1.45948049 1.1653459:1.4594805 5
18 1 y 9 1.45948049 1.75361505 1.4594805:1.753615 3
19 2 x 1 -0.61884662 -0.35602715 -.6188466:-.3560272 6
20 2 x 2 -0.35602715 -0.09320769 -.3560272:-.0932077 17
21 2 x 3 -0.09320769 0.16961178 -.0932077:.1696118 15
22 2 x 4 0.16961178 0.43243125 .1696118:.4324312 10
23 2 x 5 0.43243125 0.69525071 .4324312:.6952507 2
24 2 x 6 0.69525071 0.95807018 .6952507:.9580702 0
25 2 x 7 0.95807018 1.22088965 .9580702:1.2208896 0
26 2 x 8 1.22088965 1.48370911 1.2208896:1.4837091 0
27 2 x 9 1.48370911 1.74652858 1.4837091:1.7465286 0
28 2 y 1 -0.89359597 -0.59946141 -.893596:-.5994614 2
29 2 y 2 -0.59946141 -0.30532685 -.5994614:-.3053269 4
30 2 y 3 -0.30532685 -0.01119230 -.3053269:-.0111923 11
31 2 y 4 -0.01119230 0.28294226 -.0111923:.2829423 24
32 2 y 5 0.28294226 0.57707682 .2829423:.5770768 9
33 2 y 6 0.57707682 0.87121138 .5770768:.8712114 0
34 2 y 7 0.87121138 1.16534593 .8712114:1.1653459 0
35 2 y 8 1.16534593 1.45948049 1.1653459:1.4594805 0
36 2 y 9 1.45948049 1.75361505 1.4594805:1.753615 0
37 3 x 1 -0.61884662 -0.35602715 -.6188466:-.3560272 0
38 3 x 2 -0.35602715 -0.09320769 -.3560272:-.0932077 0
39 3 x 3 -0.09320769 0.16961178 -.0932077:.1696118 0
40 3 x 4 0.16961178 0.43243125 .1696118:.4324312 1
41 3 x 5 0.43243125 0.69525071 .4324312:.6952507 6
42 3 x 6 0.69525071 0.95807018 .6952507:.9580702 17
43 3 x 7 0.95807018 1.22088965 .9580702:1.2208896 18
44 3 x 8 1.22088965 1.48370911 1.2208896:1.4837091 4
45 3 x 9 1.48370911 1.74652858 1.4837091:1.7465286 4
46 3 y 1 -0.89359597 -0.59946141 -.893596:-.5994614 0
47 3 y 2 -0.59946141 -0.30532685 -.5994614:-.3053269 0
48 3 y 3 -0.30532685 -0.01119230 -.3053269:-.0111923 0
49 3 y 4 -0.01119230 0.28294226 -.0111923:.2829423 0
50 3 y 5 0.28294226 0.57707682 .2829423:.5770768 4
51 3 y 6 0.57707682 0.87121138 .5770768:.8712114 12
52 3 y 7 0.87121138 1.16534593 .8712114:1.1653459 26
53 3 y 8 1.16534593 1.45948049 1.1653459:1.4594805 5
54 3 y 9 1.45948049 1.75361505 1.4594805:1.753615 3
R> histogram(km.mod1)
R>
R> km.res1 <- predict(km.mod1, X, type="class", supplemental.cols = c("x","y"))
R> head(km.res1, 3)
x y CLUSTER_ID
1 -0.43646407 0.26201831 2
2 -0.02797831 0.07319952 2
3 0.11998373 -0.08638716 2
R> km.res1.local <- ore.pull(km.res1)
R> plot(data.frame(x = km.res1.local$x,
+ y = km.res1.local$y),
+ col = km.res1.local$CLUSTER_ID)
R> points(km.mod1$centers2, col = rownames(km.mod1$centers2), pch = 8, cex = 2)
R>
R> head(predict(km.mod1, X))
'2' '3' CLUSTER_ID
1 0.9992236 0.0007763706 2
2 0.9971310 0.0028690375 2
3 0.9974216 0.0025783939 2
4 0.9997335 0.0002665114 2
5 0.9917773 0.0082226599 2
6 0.9771667 0.0228333398 2
R> head(predict(km.mod1,X,type=c("class","raw"),supplemental.cols=c("x","y")),3)
'2' '3' x y CLUSTER_ID
1 0.9992236 0.0007763706 -0.43646407 0.26201831 2
2 0.9971310 0.0028690375 -0.02797831 0.07319952 2
3 0.9974216 0.0025783939 0.11998373 -0.08638716 2
R> head(predict(km.mod1,X,type="raw",supplemental.cols=c("x","y")),3)
x y '2' '3'
1 -0.43646407 0.26201831 0.9992236 0.0007763706
2 -0.02797831 0.07319952 0.9971310 0.0028690375
3 0.11998373 -0.08638716 0.9974216 0.0025783939R>
R>
R> # Text mining with ore.odmKMeans
R> title <- c('Aids in Africa: Planning for a long war',
+ 'Mars rover maneuvers for rim shot',
+ 'Mars express confirms presence of water at Mars south pole',
+ 'NASA announces major Mars rover finding',
+ 'Drug access, Asia threat in focus at AIDS summit',
+ 'NASA Mars Odyssey THEMIS image: typical crater',
+ 'Road blocks for Aids')
R> response <- c('Aids', 'Mars', 'Mars', 'Mars', 'Aids', 'Mars', 'Aids')
R>
R> KM_TEXT <- ore.push(data.frame(CUST_ID = seq(length(title)),
+ RESPONSE = response, TITLE = title))
R>
R> # Create text policy (CTXSYS.CTX_DDL privilege is required)
R> ore.exec("Begin ctx_ddl.create_policy('ESA_TXTPOL'); End;")
R>
R> # specify POLICY_NAME, MIN_DOCUMENTS, MAX_FEATURES and
R> # Text column attribute specification
R> km.mod <- ore.odmKMeans( ~ TITLE, data = KM_TEXT, num.centers = 2L,
+ odm.settings = list(ODMS_TEXT_POLICY_NAME = "ESA_TXTPOL",
+ ODMS_TEXT_MIN_DOCUMENTS = 1,
+ ODMS_TEXT_MAX_FEATURES = 3,
+ kmns_distance = "dbms_data_mining.kmns_cosine",
+ kmns_details = "kmns_details_all"),
+ ctx.settings = list(TITLE="TEXT(TOKEN_TYPE:STEM)"))
R> summary(km.mod)
Call:
ore.odmKMeans(formula = ~TITLE, data = KM_TEXT, num.centers = 2L,
odm.settings = list(ODMS_TEXT_POLICY_NAME = "ESA_TXTPOL",
ODMS_TEXT_MIN_DOCUMENTS = 1, ODMS_TEXT_MAX_FEATURES = 3,
kmns_distance = "dbms_data_mining.kmns_cosine",
kmns_details = "kmns_details_all"),
ctx.settings = list(TITLE = "TEXT(TOKEN_TYPE:STEM)"))
Settings:
value
clus.num.clusters 2
block.growth 2
conv.tolerance 0.01
details details.all
distance cosine
iterations 3
min.pct.attr.support 0.1
num.bins 10
random.seed 0
split.criterion variance
odms.missing.value.treatment odms.missing.value.auto
odms.sampling odms.sampling.disable
odms.text.max.features 3
odms.text.min.documents 1
odms.text.policy.name ESA_TXTPOL
prep.auto ON
Centers:
TITLE.MARS TITLE.NASA TITLE.ROVER TITLE.AIDS
2 0.5292307 0.7936566 0.7936566 NA
3 NA NA NA 1
R> settings(km.mod)
SETTING_NAME SETTING_VALUE SETTING_TYPE
1 ALGO_NAME ALGO_KMEANS INPUT
2 CLUS_NUM_CLUSTERS 2 INPUT
3 KMNS_BLOCK_GROWTH 2 INPUT
4 KMNS_CONV_TOLERANCE 0.01 INPUT
5 KMNS_DETAILS KMNS_DETAILS_ALL INPUT
6 KMNS_DISTANCE KMNS_COSINE INPUT
7 KMNS_ITERATIONS 3 INPUT
8 KMNS_MIN_PCT_ATTR_SUPPORT 0.1 INPUT
9 KMNS_NUM_BINS 10 INPUT
10 KMNS_RANDOM_SEED 0 DEFAULT
11 KMNS_SPLIT_CRITERION KMNS_VARIANCE INPUT
12 ODMS_MISSING_VALUE_TREATMENT ODMS_MISSING_VALUE_AUTO DEFAULT
13 ODMS_SAMPLING ODMS_SAMPLING_DISABLE DEFAULT
14 ODMS_TEXT_MAX_FEATURES 3 INPUT
15 ODMS_TEXT_MIN_DOCUMENTS 1 INPUT
16 ODMS_TEXT_POLICY_NAME ESA_TXTPOL INPUT
17 PREP_AUTO ON INPUT
R> print(predict(km.mod, KM_TEXT, supplemental.cols = "RESPONSE"), digits = 3L)
'2' '3' RESPONSE CLUSTER_ID
1 0.0213 0.9787 Aids 3
2 0.9463 0.0537 Mars 2
3 0.9325 0.0675 Mars 2
4 0.9691 0.0309 Mars 2
5 0.0213 0.9787 Aids 3
6 0.9463 0.0537 Mars 2
7 0.0213 0.9787 Aids 3
R>
R> ore.exec("Begin ctx_ddl.drop_policy('ESA_TXTPOL'); End;")