3.4 Graph Analysis Using OAAgraph
Beginning with Oracle Database 12.2 , the OAAgraph package provides an R interface to the Oracle Spatial and Graph Property Graph In-Memory Analyst (PGX) for use with Oracle R Enterprise and database tables.
About OAAgraph
PGX is an integrated set of Oracle Database functions, procedures, data types, and data models that support spatial and graph analytics. The OAAgraph package contains several graph algorithms, graph transformation operations, and graph querying capabilities.
With the OAAgraph functions in R, you can use the efficient PGX graph algorithms and representations to compute graph metrics and analysis in memory in the database. You can use the resulting data.frame objects to build models that include the graph metrics as predictors. You can use the models to score or classify data. You can then add the results to graph nodes where you can use the graph algorithms to further explore the graph or compute new metrics.
Figure 3-3 Graph Analytics and Machine Learning Interaction
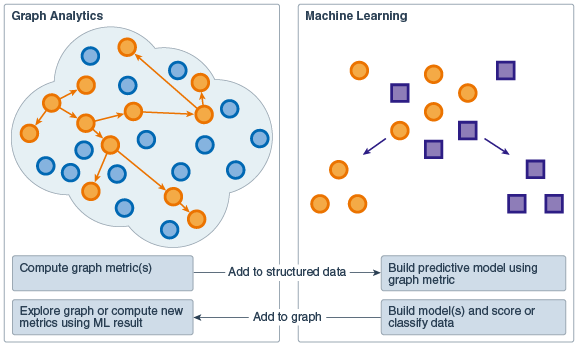
Description of "Figure 3-3 Graph Analytics and Machine Learning Interaction"
Advantages of the OAAgraph package include:
-
Access in R to the PGX in-memory graph analysis engine, which provides fast, parallel graph analysis
-
Many graph algorithms
-
Ability to query graphs and perform pattern matching
-
Integration with Oracle Big Data Spatial and Graph and Oracle R Advanced Analytics for Hadoop
Graph analysis is a methodology in data analysis in which you represent your data as a graph. Data entities become nodes and relationships become edges. You can analyze fine-grained relationships through the graph and navigate multi-hop relationships quickly without needing to repeatedly compute joins.
Two major types of graph algorithms are:
-
Computational graph analytics, which analyze an entire graph
-
Graph pattern matching, which are queries that find sub-graphs that fit relationship patterns
OAAgraph Algorithms
The algorithms in the OAAgraph package are the following:
- Ranking
-
Pagerank and variants
Vertex betweenness centrality, including approximations
Closeness centrality
Eigenvector centrality
- Path Finding
-
Dijkstra and variants
Bellman Ford and variants
Hop distance and variants
Fattest path
- Partitioning
-
Weakly and strongly connected components
Conductance and modularity
Community detection
- Recommendation
-
Twitter’s whom-to-follow
Matrix factorization
- Other
-
Breadth first search with filter
Triangle counting
Degree distribution
K-core
Adamic Adar
See Also:
For reference pages for theOAAgraph functions, see Graph Analysis Function Reference in the Oracle R Enterprise Documentation Media Library Release 1.5.1 .Example 3-77 Using OAAgraph Functions
This example uses the graph capabilities of the OAAgraph package. The example does the following:
-
Creates a graph from node and edge tables
-
Creates a graph from a snaphot in-memory representation stored in the database
-
Invokes the graph analytics algorithms
countTriangles,degree,pagerank, andadamicAdarCounting -
Uses the
oaa.cursorobject -
Cleans up in-memory graphs and database objects
library(ORE)
library(OAAgraph)
#-- Replace the values in quotation marks with the values for your database
dbHost <- "<DATABASE_HOST>"
dbUser <- "<DATABASE_USERNAME>"
dbPassword <- "<DATABASE_PASSWORD>"
dbSid <- "<DATABASE_SID>"
pgxBaseUrl <- "<PGX_BASE_URL>"
#-- Connect to the Oracle R Enterprise and PGX servers
ore.connect(host = dbHost, user = dbUser, password = dbPassword, sid = dbSid)
oaa.graphConnect(pgxBaseUrl = pgxBaseUrl, dbHost = dbHost,
dbSid = dbSid, dbUser = dbUser, dbPassword = dbPassword)
#-- Create the node table in Oracle Database
VID <- c(1, 2, 3, 4, 5)
NP1 <- c("node1", "node2", "node3", "node4", "node5")
NP2 <- c(111.11, 222.22, 333.33, 444.44, 555.55)
NP3 <- c(1, 2, 3, 4, 5)
nodes <- data.frame(VID, NP1, NP2, NP3)
ore.drop(table = "MY_NODES")
ore.create(nodes, table = "MY_NODES")
#-- Create the edge table in Oracle Database
EID <- c(1, 2, 3, 4, 5)
SVID <- c(1, 3, 3, 2, 4)
DVID <- c(2, 1, 4, 3, 2)
EP1 <- c("edge1", "edge2", "edge3", "edge4", "edge5")
EL <- c("label1", "label2", "label3", "label4", "label5")
edges <- data.frame(EID, SVID, DVID, EP1, EL)
ore.drop(table = "MY_EDGES")
ore.create(edges, table = "MY_EDGES")
#-- Verify that the tables exist as ore.frame objects
ore.ls()
#-- Create a graph in PGX from the node and edge tables in the database
graph <- oaa.graph(MY_EDGES, MY_NODES, "myPgxGraph")
names(graph, "nodes")
names(graph, "edges")
#-- See the result of the countTriangles function, which gives an
#-- overview of the number of connections between nodes in neighborhoods
countTriangles(graph, sortVerticesByDegree=FALSE)
#-- See the results from degree algorithm variants, note the graph nodes
#-- are augmented with new properties as indicated by the 'name' argument
degree(graph, name = "OutDegree")
degree(graph, name = "InDegree", variant = "in")
degree(graph, name = "InOutDegree", variant = "all")
#-- Create a cursor including the degree properties
cursor <- oaa.cursor(graph, c("OutDegree", "InOutDegree", "InDegree"), "nodes")
oaa.next(cursor, 5)
#-- Create a cursor over the degree properties using
#-- the PGX SQL-like query language PGQL
cursor <- oaa.cursor(graph,
query = "select n.OutDegree, n.InOutDegree, n.InDegree
where (n) order by n.OutDegree desc")
#-- View the first 5 entries from the cursor
oaa.next(cursor, 5)
#-- See results from the pagerank algorithm
pagerankCursor <- pagerank(graph, 0.085, 0.1, 100)
oaa.next(pagerankCursor, 5)
#-- Create a cursor over the pagerank property using PGQL
cursor <- oaa.cursor(graph,
query = "select n.pagerank where (n)
order by n.pagerank desc")
oaa.next(cursor, 5)
#-- You can create a cursor using the R interface as well
cursor <- oaa.cursor(graph, "pagerank", ordering = "desc")
oaa.next(cursor, 5)
#-- Compute the adamic adar index for edges
topEdges <- adamicAdarCounting(graph)
oaa.next(topEdges)
#-- List any graph snapshots available
oaa.graphSnapshotList()
#-- Export a binary snapshot of the whole graph into Oracle Database
#-- and view the listing again
oaa.graphSnapshotPersist(graph, nodeProperties = TRUE, edgeProperties = TRUE)
oaa.graphSnapshotList()
#-- Read the snapshot back into memory
graph2 <- oaa.graphSnapshot("myPgxGraph")
#-- Export the graph nodes and specific node properties from memory
#-- into a database table
oaa.create(graph2, nodeTableName = "RANKED_NODES", nodeProperties = TRUE)
#-- Export both nodes and edges as tables from memory into the database,
#-- but only export the pagerank node property
oaa.create(graph2, nodeTableName = "RANKED_GRAPH_N",
nodeProperties = c("NP1", "pagerank"),
edgeTableName = "RANKED_GRAPH_E")
#-- Export the graph edges and their properties from memory into a database table
oaa.create(graph2, edgeTableName = "RANKED_EDGES", edgeProperties = TRUE)
#-- Free the graphs at the PGX server
oaa.rm(graph)
oaa.rm(graph2)
#-- Clean up the tables created by this example
ore.drop("MY_NODES")
ore.drop("MY_EDGES")
ore.drop("RANKED_NODES")
ore.drop("RANKED_GRAPH_N")
ore.drop("RANKED_GRAPH_E")
ore.drop("RANKED_EDGES")
oaa.dropSnapshots("myPgxGraph")
Listing for This Example
R> library(ORE)
R> library(OAAgraph)
R>
R> #-- Replace the values in quotation marks with the values for your database
R> dbHost <- "<DATABASE_HOST>"
R> dbUser <- "<DATABASE_USERNAME>"
R> dbPassword <- "<DATABASE_PASSWORD>"
R> dbSid <- "<DATABASE_SID>"
R> pgxBaseUrl <- "<PGX_BASE_URL>"
R>
R> #-- Connect to the Oracle R Enterprise and PGX servers
R> ore.connect(host = dbHost, user = dbUser, password = dbPassword, sid = dbSid)
R> oaa.graphConnect(pgxBaseUrl = pgxBaseUrl, dbHost = dbHost,
+ dbSid = dbSid, dbUser = dbUser, dbPassword = dbPassword)
R>
R> #-- Create the node table in Oracle Database
R>
R> VID <- c(1, 2, 3, 4, 5)
R> NP1 <- c("node1", "node2", "node3", "node4", "node5")
R> NP2 <- c(111.11, 222.22, 333.33, 444.44, 555.55)
R> NP3 <- c(1, 2, 3, 4, 5)
R>
R> nodes <- data.frame(VID, NP1, NP2, NP3)
R> ore.drop(table = "MY_NODES")
R> ore.create(nodes, table = "MY_NODES")
R>
R> #-- Create the edge table in Oracle Database
R>
R> EID <- c(1, 2, 3, 4, 5)
R> SVID <- c(1, 3, 3, 2, 4)
R> DVID <- c(2, 1, 4, 3, 2)
R> EP1 <- c("edge1", "edge2", "edge3", "edge4", "edge5")
R> EL <- c("label1", "label2", "label3", "label4", "label5")
R>
R> edges <- data.frame(EID, SVID, DVID, EP1, EL)
R>
R> ore.drop(table = "MY_EDGES")
R> ore.create(edges, table = "MY_EDGES")
R>
R> #-- Verify that the tables exist as ore.frame objects
R>
R> ore.ls()
[1] "ASSIGN_EDGES_SUBSET" "ASSIGN_NODES_SUBSET" "CALL_EDGES"
[4] "DF_EDGES_140317215226" "DF_EDGES_150317002703" "DF_NODES_140317215226"
[7] "DF_NODES_150317002703" "EDGES" "EDGES_KEY"
[10] "EDGES_T" "MY_EDGES" "MY_NODES"
[13] "MY_NODES1" "N_H5855" "NODES"
[16] "NODES2_T" "NODES30174128" "NODES30174506"
[19] "NODES30174740" "NODES_KEY" "NODES_T"
[22] "nyc20m" "PERSON_NODES" "PERSON_PAGERANK_NODES"
[25] "SCCE" "SCCN" "SUPERHERO_DATA"
[28] "SUPERHERO_DATA2" "SUPERHERO_EDGES" "SUPERHERO_IGNORE"
[31] "SUPERHERO_INFORMATION" "SUPERHERO_NODES" "SUPERHERO_VALUES"
[34] "TABLE1" "TABLE2" "TEMP_EDGES"
[37] "TEMP_NODES" "TMPN"
R>
R> #-- Create a graph in PGX from the node and edge tables in the database
R>
R> graph <- oaa.graph(MY_EDGES, MY_NODES, "myPgxGraph")
R> names(graph, "nodes")
[1] "NP1" "NP3" "NP2"
R> names(graph, "edges")
[1] "EP1"
R>
R> #-- See the result of the countTriangles function, which gives an
R> #-- overview of the number of connections between nodes in neighborhoods
R>
R> countTriangles(graph, sortVerticesByDegree=FALSE)
[1] 2
R>
R> #-- See the results from degree algorithm variants; note the graph nodes
R> #-- are augmented with new properties as indicated by the 'name' argument
R>
R> degree(graph, name = "OutDegree")
oaa.cursor over: ID, OutDegree
position: 0
size: 5
R> degree(graph, name = "InDegree", variant = "in")
oaa.cursor over: ID, InDegree
position: 0
size: 5
R> degree(graph, name = "InOutDegree", variant = "all")
oaa.cursor over: ID, InOutDegree
position: 0
size: 5
R>
R> #-- Create a cursor including the degree properties
R>
R> cursor <- oaa.cursor(graph, c("OutDegree", "InOutDegree", "InDegree"), "nodes")
R> oaa.next(cursor, 5)
OutDegree InOutDegree InDegree
1 1 2 1
2 1 3 2
3 2 3 1
4 1 2 1
5 0 0 0
R>
R> #-- Create a cursor over the degree properties using
R> #-- the PGX SQL-like query language PGQL
R>
R> cursor <- oaa.cursor(graph,
+ query = "select n.OutDegree, n.InOutDegree, n.InDegree
+ where (n) order by n.OutDegree desc")
R> #-- View the first 5 entries from the cursor
R>
R> oaa.next(cursor, 5)
n.OutDegree n.InOutDegree n.InDegree
1 2 3 1
2 1 3 2
3 1 2 1
4 1 2 1
5 0 0 0
R>
R> #-- See the results from the pagerank algorithm
R>
R> pagerankCursor <- pagerank(graph, 0.085, 0.1, 100)
R> oaa.next(pagerankCursor, 5)
pagerank
2 0.22
3 0.20
1 0.19
4 0.19
5 0.18
R>
R> #-- Create a cursor over the pagerank property using PGQL
R>
R> cursor <- oaa.cursor(graph,
+ query = "select n.pagerank where (n)
+ order by n.pagerank descR> ")
R>
R> oaa.next(cursor, 5)
n.pagerank
1 0.22
2 0.20
3 0.19
4 0.19
5 0.18
R>
R> #-- You can create a cursor using the R interface as well
R>
R> cursor <- oaa.cursor(graph, "pagerank", ordering = "desc")
R>
R> oaa.next(cursor, 5)
pagerank
1 0.19
2 0.22
3 0.20
4 0.19
5 0.18
R>
R> #-- Compute the adamic adar index for edges
R>
R> topEdges <- adamicAdarCounting(graph)
R> oaa.next(topEdges)
adamic_adar
0 0
1 0
2 0
3 0
4 0
R>
R> #-- List any graph snapshots available
R>
R> oaa.graphSnapshotList()
[1] "ANONYMOUS_GRAPH_1" "CONNECTIONS" "EXAMPLE_GRAPH"
[4] "GRAPH1" "GRAPH_EXPORT_LABELED" "G_160317161147"
[7] "G_160317201914" "MYAWESOMEGRAPH" "MYEXAMPLEGRAPH"
[10] "MY_GRAPH1" "SAMPLE" "SAMPLE_GRAPH"
[13] "SF" "SF_MUTATION"
R>
R> #-- Export a binary snapshot of the whole graph into Oracle Database
R> #-- and view the listing again
R>
R> oaa.graphSnapshotPersist(graph, nodeProperties = TRUE, edgeProperties = TRUE)
R> oaa.graphSnapshotList()
[1] "ANONYMOUS_GRAPH_1" "CONNECTIONS" "EXAMPLE_GRAPH"
[4] "GRAPH1" "GRAPH_EXPORT_LABELED" "G_160317161147"
[7] "G_160317201914" "MYAWESOMEGRAPH" "MYEXAMPLEGRAPH"
[10] "MYPGXGRAPH" "MY_GRAPH1" "SAMPLE"
[13] "SAMPLE_GRAPH" "SF" "SF_MUTATION"
R>
R> #-- Read the snapshot back into memory
R>
R> graph2 <- oaa.graphSnapshot("myPgxGraph")
R>
R> #-- Export the graph nodes and specific node properties from memory
R> #-- into a database table
R>
R> oaa.create(graph2, nodeTableName = "RANKED_NODES", nodeProperties = TRUE)
R>
R> #-- Export both nodes and edges as tables from memory into the database,
R> #-- but only export the pagerank node property
R>
R> oaa.create(graph2, nodeTableName = "RANKED_GRAPH_N",
+ nodeProperties = c("NP1", "pagerank"),
+ edgeTableName = "RANKED_GRAPH_E")
R>
R> #-- Export the graph edges and their properties from memory into a database table
R>
R> oaa.create(graph2, edgeTableName = "RANKED_EDGES", edgeProperties = TRUE)
R>
R> #-- Free the graphs at the PGX server
R>
R> oaa.rm(graph)
R> oaa.rm(graph2)
R>
R> #-- Clean up the tables created by this example
R>
R> ore.drop("MY_NODES")
R> ore.drop("MY_EDGES")
R> ore.drop("RANKED_NODES")
R> ore.drop("RANKED_GRAPH_N")
R> ore.drop("RANKED_GRAPH_E")
R> ore.drop("RANKED_EDGES")
R>
R> oaa.dropSnapshots("myPgxGraph")