3.1 Ontologies
An ontology is a shared conceptualization of knowledge in a particular domain.
It consists of a collection of classes, properties, and optionally instances. Classes are typically related by class hierarchy (subclass/ superclass relationship). Similarly, the properties can be related by property hierarchy (subproperty/ superproperty relationship). Properties can be symmetric or transitive, or both. Properties can also have domain, ranges, and cardinality constraints specified for them.
RDFS-based ontologies only allow specification of class hierarchies, property hierarchies, instanceOf relationships, and a domain and a range for properties.
OWL ontologies build on RDFS-based ontologies by additionally allowing specification of property characteristics. OWL ontologies can be further classified as OWL-Lite, OWL-DL, and OWL Full. OWL-Lite restricts the cardinality minimum and maximum values to 0 or 1. OWL-DL relaxes this restriction by allowing minimum and maximum values. OWL Full allows instances to be also defined as a class, which is not allowed in OWL-DL and OWL-Lite ontologies.
Supported OWL Subsets describes OWL capabilities that are supported and not supported with RDF data.
Parent topic: OWL Concepts
3.1.1 Example: Disease Ontology
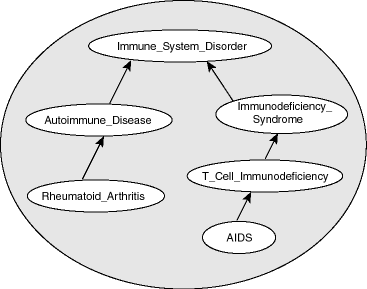
Figure 3-1 shows part of a disease ontology, which describes the classes and properties related to certain diseases. One requirement is to have a PATIENTS data table with a column named DIAGNOSIS, which must contain a value from the Diseases_and_Disorders class hierarchy.
In the disease ontology shown in Figure 3-1, the diagnosis Immune_System_Disorder includes two subclasses, Autoimmune_Disease and Immunodeficiency_Syndrome. The Autoimmune_Disease diagnosis includes the subclass Rheumatoid_Arthritis; and the Immunodeficiency_Syndrome diagnosis includes the subclass T_Cell_Immunodeficiency, which includes the subclass AIDS.
The data in the PATIENTS table might include the PATIENT_ID and DIAGNOSIS column values shown in Table 3-1.
Table 3-1 PATIENTS Table Example Data
| PATIENT_ID | DIAGNOSIS |
|---|---|
|
1234 |
Rheumatoid_Arthritis |
|
2345 |
Immunodeficiency_Syndrome |
|
3456 |
AIDS |
To query ontologies, you can use the SEM_MATCH table function or the SEM_RELATED operator and its ancillary operators.
Related Topics
Parent topic: Ontologies
3.1.2 Supported OWL Subsets
This section describes OWL vocabulary subsets that are supported.
Oracle AI Database supports the RDFS++, OWLSIF, and OWLPrime vocabularies, which have increasing expressivity, as well as OWL 2 RL. Each supported vocabulary has a corresponding rulebase; however, these rulebases do not need to be populated because the underlying inference rules of these three vocabularies are internally implemented. The supported vocabularies are as follows:
-
RDFS++: A minimal extension to RDFS; which is RDFS plus
owl:sameAsandowl:InverseFunctionalProperty. -
OWLSIF: OWL with IF Semantic, with the vocabulary and semantics proposed for pD* semantics in Completeness, decidability and complexity of entailment for RDF Schema and a semantic extension involving the OWL vocabulary, by H.J. Horst, Journal of Web Semantics 3, 2 (2005), 79–115.
-
OWLPrime: The following OWL capabilities:
-
Basics: class, subclass, property, subproperty, domain, range, type
-
Property characteristics: transitive, symmetric, functional, inverse functional, inverse
-
Class comparisons: equivalence, disjointness
-
Property comparisons: equivalence
-
Individual comparisons: same, different
-
Class expressions: complement
-
Property restrictions:
hasValue,someValuesFrom,allValuesFromAs with pD*, the supported semantics for these value restrictions are only intensional (IF semantics).
-
-
OWL 2 RL: Described in the "OWL 2 RL" section of the W3C OWL 2 Web Ontology Language Profiles recommendation (
http://www.w3.org/TR/owl2-profiles/#OWL_2_RL) as: "The OWL 2 RL profile is aimed at applications that require scalable reasoning without sacrificing too much expressive power. It is designed to accommodate both OWL 2 applications that can trade the full expressivity of the language for efficiency, and RDF(S) applications that need some added expressivity from OWL 2."The system-defined rulebase
OWL2RLsupports all the standard production rules defined for OWL 2 RL. As withOWLPRIME, users will not see any rules in this OWL2RL rulebase. The rulebaseOWL2RLwill be created automatically if it does not already exist.The following code excerpt uses the OWL2RL rulebase:
EXECUTE sem_apis.create_rdf_graph('m1',NULL, NULL, network_owner=>'RDFUSER', network_name=>'NET1'); -- Insert data into RDF graph M1. Details omitted ... -- Now run inference using the OWL2RL rulebase EXECUTE sem_apis.create_inferred_graph('m1_inf',sem_models('m1'),sem_rulebases('owl2rl'),network_owner=>'RDFUSER',network_name=>'NET1');Note that inference-related optimizations, such as parallel inference and RAW8, are all applicable when the
OWL2RLrulebase is used. -
OWL 2 EL: Described in the "OWL 2 EL" section of the W3C OWL 2 Web Ontology Language Profiles recommendation (
http://www.w3.org/TR/owl2-profiles/#OWL_2_EL) as: "The OWL 2 EL profile is designed as a subset of OWL 2 that:- is particularly suitable for applications employing ontologies that define very large numbers of classes and/or properties,
- captures the expressive power used by many such ontologies, and
- for which ontology consistency, class expression subsumption, and instance checking can be decided in polynomial time."
A prime example of OWL 2 EL ontology is the biomedical ontology SNOMED Clinical Terms (SNOMED CT). For information about SNOMED CT, see:
http://www.ihtsdo.org/snomed-ct/The system-defined rulebase
OWL2ELsupports the EL syntax.As with
OWLPRIMEandOWL2RL, users will not see any rules in thisOWL2ELrulebase, and theOWL2ELrulebase will be created automatically if it does not already exist.The following code excerpt uses the
OWL2ELrulebase against the well known SNOMED ontology:EXECUTE sem_apis.create_rdf_graph('snomed',NULL,NULL,network_owner=>'RDFUSER',network_name=>'NET1') compress; -- Insert data into RDF graph SNOMED. Details omitted ... -- Now run inference using the OWL2EL rulebase EXECUTE sem_apis.create_inferred_graph('snomed_inf',sem_models('snomed'),sem_rulebases('owl2el'),network_owner=>'RDFUSER',network_name=>'NET1');Note that the
OWL2ELrulebase support does not include reflexive object properties (ReflexiveObjectProperty) simply because a reflexive object property will link every individual with itself, which would probably cause an unnecessary and costly expansion of the inference graph.
Table 3-2 lists the RDFS/OWL vocabulary constructs included in each supported rulebase.
Table 3-2 RDFS/OWL Vocabulary Constructs Included in Each Supported Rulebase
| Rulebase Name | RDFS/OWL Constructs Included |
|---|---|
|
RDFS++ |
all RDFS vocabulary constructs owl:InverseFunctionalProperty owl:sameAs |
|
OWLSIF |
all RDFS vocabulary constructs owl:FunctionalProperty owl:InverseFunctionalProperty owl:SymmetricProperty owl:TransitiveProperty owl:sameAs owl:inverseOf owl:equivalentClass owl:equivalentProperty owl:hasValue owl:someValuesFrom owl:allValuesFrom |
|
OWLPrime |
rdfs:subClassOf rdfs:subPropertyOf rdfs:domain rdfs:range owl:FunctionalProperty owl:InverseFunctionalProperty owl:SymmetricProperty owl:TransitiveProperty owl:sameAs owl:inverseOf owl:equivalentClass owl:equivalentProperty owl:hasValue owl:someValuesFrom owl:allValuesFrom owl:differentFrom owl:disjointWith owl:complementOf |
|
OWL2RL |
(As described in |
|
OWL2EL |
(As described in |
Parent topic: Ontologies