Retrieve Variant Data in VCF Format and Specimen Metadata for a Patient Cohort
This use case shows you how to retrieve variant data in VCF format for a patient cohort along with the specimen metadata.
Use Case
Retrieve patients for a clinical criteria and export variant data in VCF format along with specimen metadata from genomic and clinical side. Also save the patients as a cohort list.
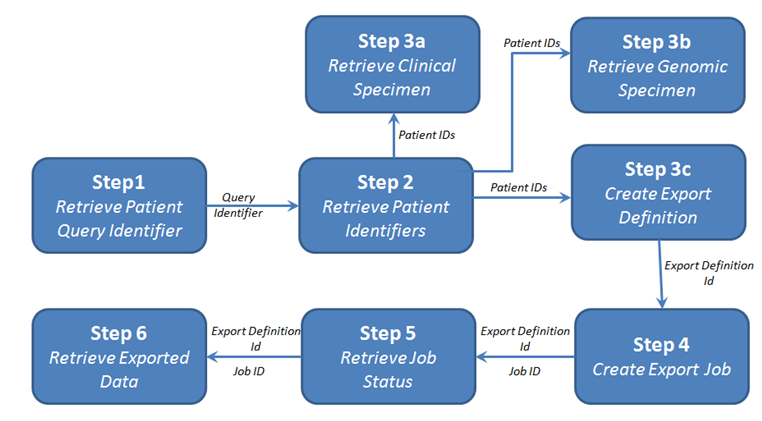
Step 1: Retrieve Patient Query Identifiers
In this step, you retrieve a list of patient IDs conforming to the criteria defined in the following request. For details, see the Invoke Patient Query endpoint.
Request URL
http://localhost:7010/ohfapi/cg/3.3/patientapi/patients/queries
Request Type
POST
Request Body
{
"queryCriteria":{
"allCriteriaList":[
{
"type":"ConsentCriteria",
"consentTypeCodeList":[
"CNSNT_TYP_SPCMN_CNSNT"
]
},
{
"type": "DiagnosisCriteria",
"diagnosisCodeList": [
"N179"
]
},
{
"type": "DemographicsCriteria",
"genderNameList": [
"FEMALE"
]
},
{
"type": "MedicationCriteria",
"medicationCodeList": [
"1114470"
],
"medicationStartDateRange": {
"value": "01/01/2020",
"op": {
"code": "COMP_OP_GE"
}
},
"medicationEndDateRange": {
"value": "01/31/2020",
"op": {
"code": "COMP_OP_LE"
}
},
"medicationDosageRange": {
"value": 88,
"op": {
"code": "COMP_OP_EQ"
}
},
"dosageUOMCodeList": [
"1001"
]
}
]
}
}
JSON Response
[ {
"rel" : "status",
"href" : "/ohfapi/cg/v3.3/patientapi/patients/queries/17503"
}, {
"rel" : "result",
"href" : "/ohfapi/cg/v3.3/patientapi/patients/queries/17503/data"
} ]
Step 2: Retrieve Patient Identifiers for Query Identifier
This API uses the Reference URL retrieved in Step 1. For details, see the Retrieve Patient Query Results for a Query Identifierendpoint.
Request URL
http://localhost:7010/ohfapi/cg/v3.3/patientapi/patients/queries/17503/data
Request Type
GET
JSON Response
{
"items" : [ {
"patientId" : 823,
"patientIdentifier" : "7"
}, {
"patientId" : 838,
"patientIdentifier" : "2"
}, {
"patientId" : 839,
"patientIdentifier" : "91"
} ],
"count" : 3,
"hasMore" : false,
"links" : [ {
"rel" : "self",
"href" : "http://localhost:7010/ohfapi/cg/v3.3/patientapi/patients/queries/17503/data"
} ]
}
Step 3a: Retrieve Clinical Specimen Metadata for Specimen ID
This API uses the Specimen ID (823) retrieved in Step 2 to fetch the Clinical Specimens. For details, see the Retrieve Clinical Specimen Metadata for a Specimen Identifier endpoint.
Request URL
http://localhost:7010/ohfapi/cg/v3.3/patientapi/clinicalspecimens/823/metadata
Request Type
GET
JSON Response
{
"resourceType" : "SpecimenMetadata",
"items" : [ {
"identifier" : {
"system" : "CDM",
"value" : "10056"
},
"metadata" : {
"attrName" : "CAPTURE_KIT",
"attrDesc" : "Capture Kit",
"strValue" : " Agilent SS Custom Capture",
"strValueUpper" : " AGILENT SS CUSTOM CAPTURE"
},
"specimenReference" : {
"reference" : "/ohfapi/cg/v3.3/patientapi/clinicalspecimens/1080",
"display" : "589(SVCPRV 12)"
},
"specimenVendorNumber" : "SVCPRV 12",
"accessionIdentifier" : "589"
}, {
"identifier" : {
"system" : "CDM",
"value" : "10054"
},
"metadata" : {
"attrName" : "CUSTOM_INDEX",
"attrDesc" : "Custom Index",
"strValue" : "IGCCAAT",
"strValueUpper" : "IGCCAAT"
},
"specimenReference" : {
"reference" : "/ohfapi/cg/v3.3/patientapi/clinicalspecimens/1080",
"display" : "589(SVCPRV 12)"
},
"specimenVendorNumber" : "SVCPRV 12",
"accessionIdentifier" : "589"
}
],
"count" : 2,
"hasMore" : false,
"links" : [ {
"rel" : "self",
"href" : "http://localhost:7010/ohfapi/cg/v3.3/patientapi/clinicalspecimens/823/metadata"
} ]
}
Step 3b: Retrieve Genomic Specimens for Specimen IDs
This API uses the Specimen ID (823) retrieved in Step 2 to fetch the Genomic Specimens. For details, see the Retrieve Genomic Specimen Metadata for Specimen Identifier endpoint.
Request URL
http://localhost:7010/ohfapi/cg/v3.3/patientapi/genomicspecimens/823/metadata
Request Type
GET
JSON Response
{
"resourceType": "GenomicSpecimenMetadata",
"items": [
{
"categoryName": "ANALYSIS",
"metadata": [
{
"tag": "DATATYPE",
"strValue": "variant"
},
{
"tag": "GENOMEBUILD",
"strValue": "GRCh37"
},
{
"tag": "ANALYSISSYS",
"strValue": "NGS"
},
{
"tag": "DELETEEXISTINGRESULT",
"strValue": "Y"
},
{
"tag": "ANALYSISID",
"strValue": "GENOME_GPS"
},
{
"tag": "ANALYSISVERSION",
"strValue": "2.0.2"
},
{
"tag": "ANALYSISDATE",
"strValue": "03/04/2015 02:43",
"dateValue": "03/04/2015"
},
{
"tag": "DATAFILE",
"strValue": "dataFiles/gvcf/14-001385_1409700144.gvcf.gz"
}
]
}
],
"count": 1,
"hasMore": false,
"links": [
{
"rel": "self",
"href": "http://localhost:7010/ohfapi/cg/v3.3/patientapi/genomicspecimens/823/metadata"
}
]
}
Step 3c: Create Export Definition
After retrieving the patient IDs in step 2, you can export the variants from these patients in VCF format. To do this, create the export definition with the patients and other details such as exporting variants from a gene (PTEN) as described in the following request. This API uses the Patient IDs retrieved in Step 2 in the request body. For details, see the Create Export Definition endpoint.
Request URL
http://localhost:7010/ohfapi/cg/v3.3/exports
Request Type
POST
Request Body
{
"context": "PATIENT",
"geneHugoNames":["PTEN"],
"alignmentId": 1,
"patientSubjectId": ["823","838","839"],
"serviceProviderIdentifier": "vcf"
}
JSON Response
{
"exportDefId" : {
"system" : "ODB",
"value" : "2032"
},
"link" : {
"rel" : "SUBMITJOB",
"href" : "/ohfapi/cg/v3.3/exports/2032/jobs"
}
}
Step 4: Create Export Job for Export Definition
After successfully creating the export definition, create the Job using this definition. You can check the status of the job using the URL in the response. This API uses the Export Definition ID retrieved in Step 3. For details, see the Create Export Job for an Export Definition endpoint.
Request URL
http://localhost:7010/ohfapi/cg/v3.3/exports/2032/jobs
Request Type
POST
JSON Response
{
"exportId" : "2032",
"link" : {
"rel" : "GETSTATUS",
"href" : "/ohfapi/cg/v3.3/exports/2032/jobs/6001"
}
}
Step 5: Retrieve Job Status
This API retrieved the job status using the Job ID retrieved in Step 4. For details, see the Retrieve Job Status for an Export Identifier and Job Identifier endpoint.
Request URL
http://localhost:7010/ohfapi/cg/v3.3/exports/2032/jobs/6001
Request Type
GET
JSON Response
{
"exportId" : 2032,
"jobId" : 6001,
"status" : "SUCCEEDED",
"links" : [ {
"rel" : "GETSTATUS",
"href" : "/ohfapi/cg/v3.3/exports/2032/jobs/6001"
}, {
"rel" : "GETDATA",
"href" : "/ohfapi/cg/v3.3/exports/2032/jobs/6001/data"
} ]
}
Step 6: Retrieve Exported Data for the Job
After the export job is completed, download the VCF file using the URL given in the response. For details, see the Retrieve Exported Data for a Job endpoint.
To retrieve the exported data:
-
Enter the following path in the browser:
http://localhost:7010/ohfapi/cg/v3.3/exports/2032/jobs/6001/data
Note:
This request does not work in Postman Client or other tools. You must use a browser. -
Save the resulting zip file.
-
Unzip and view the contents to see the exported data.