| Oracle® Health Sciences Omics Data Bank Programmer's Guide Release 3.0.2.1 E35680-12 |
|
|
PDF · Mobi · ePub |
| Oracle® Health Sciences Omics Data Bank Programmer's Guide Release 3.0.2.1 E35680-12 |
|
|
PDF · Mobi · ePub |
This chapter contains the following topics:
Oracle Health Sciences Omics Data Bank (ODB) consists of two groups of tables. One set of tables, called the reference set, provides the genomic feature metadata required to link specimen sample results to specific regions of the genome, gene definitions, pathway or protein definitions. The second set of tables, called the result set, captures the specimen sample genomic results, and links each result to an object in the reference model. Each specimen sample is linked back to the patient and (or) subject. The patient or subject link is accomplished by linking ODB with Cohort Data Model (part of Oracle Health Sciences Cohort Explorer).
Omics Data Bank (ODB) consists of the data model and loaders. It does not include any front-end user interfaces and contains only command line APIs. A set of scripts, included with the model, can be used to load reference genomic data from specific sources and several types of result data from a limited set of formats.
The model is intended to handle very large amounts of data (of the order of terabytes and more). To gauge the scale of the data, one should consider that the human genome has around 3 billion bases in each strand and the number of genes that produce proteins is around 23,000. Each gene has many different variants and attributes that are described in detail. This holds true for each protein, gene component, pathway and so on, thus a lot of supporting reference data is loaded for each gene, protein or pathway. The data cannot merely be reloaded to maintain an organized table, as is the case with other tables that use ETL processes to load data. One approach is to use Index Organized tables since data can be added to the reference model as more reference data is discovered for each gene.
ODB 3.0 can handle multiple versions of reference data in the same instance, for example GRCh36, GRCh37. You can load multiple versions of each reference data, even the same version multiple times, and link the results to all copies of a specific reference version based on the requirement. For example, user with sequencing data mapped to GRCh36.p7 links the results to GRCh36.p7 reference data loaded from ENSEMBL. The multiple reference version support is extended to ENSEMBL, SwissProt, HUGO, Pathway, SIFT or PolyPhen, COSMIC, HGMD, ADF, and probe loader.
The ODB model can also handle multiple species concurrently, both on the reference and result side of the data model.
Reference data is loaded from the following distinct sources:
The genomic information with corresponding gene data is loaded from EMBL files which are stored online in the Ensembl database (http://www.ensembl.org/). Ensembl is a joint project of the European Bioinformatics Institute and the Wellcome Trust Sanger Institute. This online database maintains references to other online database projects (dbSNP, NCBI, Cosmic, and so on) and provides references to each of these databases. The model loads this cross reference information, including known variation data, to let queries use specific database references, if needed. Files in the genuine EMBL format from other sources can be usually loaded in the same way (However, their successful loading cannot be guaranteed). Each release of Ensembl data is treated as a separate 'DNA' reference version. Current and alternate releases are available at their FTP publication repository. Each newly loaded 'DNA' reference version should be given a new label. When loading variants or proteins, each given variant or protein record is linked to a specific version of the DNA reference.
The second source is the online SwissProt database (http://www.ebi.ac.uk/uniprot/) from which the model obtains protein information. This database project is also a consortium of various groups including the European Bioinformatics Institute. An FTP link to the current release of the SwissProt file is provided on their download page. Older SwissProt releases are stored as large compressed files in their FTP repository.
The third source is the HUGO Gene Nomenclature Committee (http://www.genenames.org/). This source provides reference seed information required to identify human gene locations annotation only. The HUGO gene names are required to find various cross references and the correct chromosome number for each gene. The HUGO Gene Nomenclature Committee is the authoritative group for all gene names. Since the HGNC HUGO dataset is continuously updated, it does not have an archive of older versions or releases. Any dataset taken, is as current as the date it was retrieved.
The fourth reference source is PathwayCommons (http://www.pathwaycommons.org/pc/), which is used as the source for published pathways and proteins or genes participating in each pathway. The coverage of pathway as a reference is minimal. Pathway provides a list of current and previous releases of the dataset to be retrieved from its download page.
The fifth reference source is Human Genome Mutation Database (HGMD) (http://www.hgmd.cf.ac.uk/ac/index.php) This reference source currently provides information on inherited genomic variants, as well as information connecting inherited mutations and genes with human diseases and pharmacological effects. The latter is obtained from HGMD's commercial partner, BioBase International.
The sixth reference source is Catalogue of Somatic Mutations in Cancer (COSMIC) (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic), which is a source of information related to human cancers including somatic mutation, sample annotation, and publication ID links. This provides datasets on coding and non-coding variants, and corresponding annotative information for each, consisting of sample origin, histology, publication, and genomic data.
In general, the above files use similar features to represent data. The EMBL format is a flat file representation, which provides an easy mechanism to parse and store data in a separate database structure. Both Ensembl and SwissProt have native schemas that can be downloaded. However, these schemas have 3NF structures, which are cumbersome to coerce into a star schema model. The ODB model does not copy the source schema structures in any way. In addition, there are a lot of extra objects in the native schemas that are not necessary for the type of queries needed for the ODB requirements and these objects are omitted in the ODB schema.
Most of these online databases let you download complete references, or specific references for sections of the genome. Since some customers may only need some genes or proteins, and some may not need any protein information at all, the model permits any combination of specific data to be loaded and updated. Ensembl and SwissProt databases are maintained in an additive manner, so that new data is added on top of the existing data. This lets the ODB reference data be expanded as required by the customer. The HUGO, PathwayCommons, and HGMD databases do not keep older versions but provide only the latest versions for download.
In ODB, the data model handles two main types of genetic results—gene expression and sequencing. Gene Expression experiments capture information on how effectively certain genes respond to various conditions, or how they differentially express under different conditions. For sequencing results, while there are many different types of sequencing techniques, the net effect is to record all of the variants which include SNPs, small indels, large structural variations, structural re-arrangements, and also non-variant information detected for each sample being tested, copy number variation and other related features.
The model is designed to facilitate easy querying of all the above result types in a single SQL statement across multiple versions of references or within the specific reference version.
ODB contains two sets of tables:
Reference data tables
Result data tables
Each set of tables comes with a set of loading scripts to load data into these tables. The reference loaders write to the reference tables, while the result loaders write to result tables, and link results to reference. However, there is one exception, the W_EHA_VARIANT reference table, where the sequencing result loaders enable you to report on any novel variants by writing to this table with any new variants found. A dedicated procedure, invoked by the result loader, reports on any novel variants.
Figure 1-1 shows how the reference tables link to create the ODB reference. Only table names are shown in the figure.
Figure 1-1 Reference Data Logical Model (Core Tables Only)
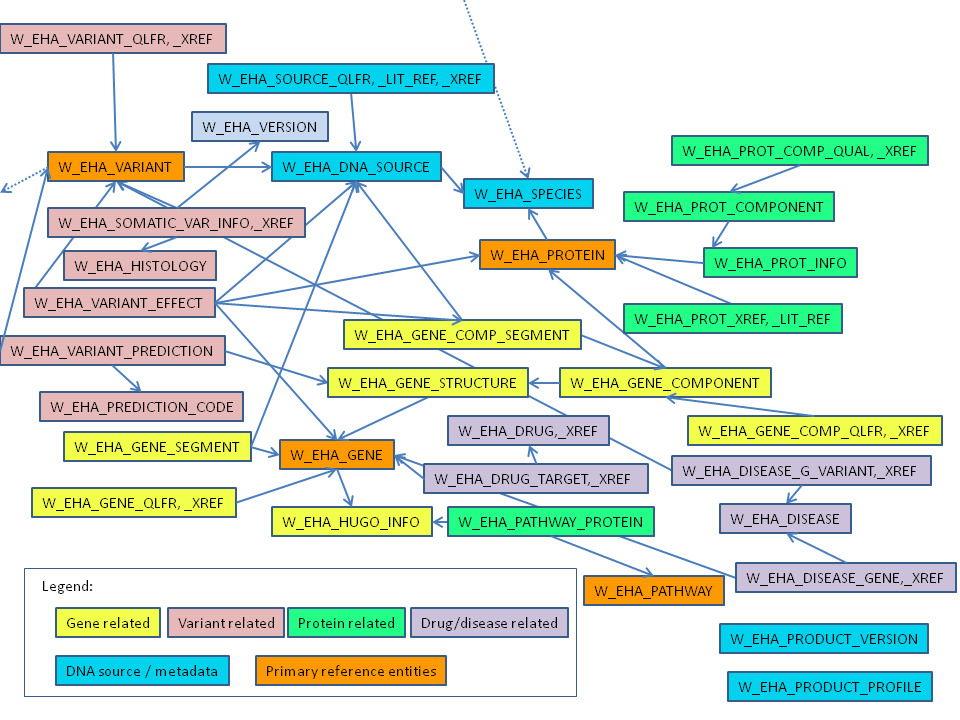
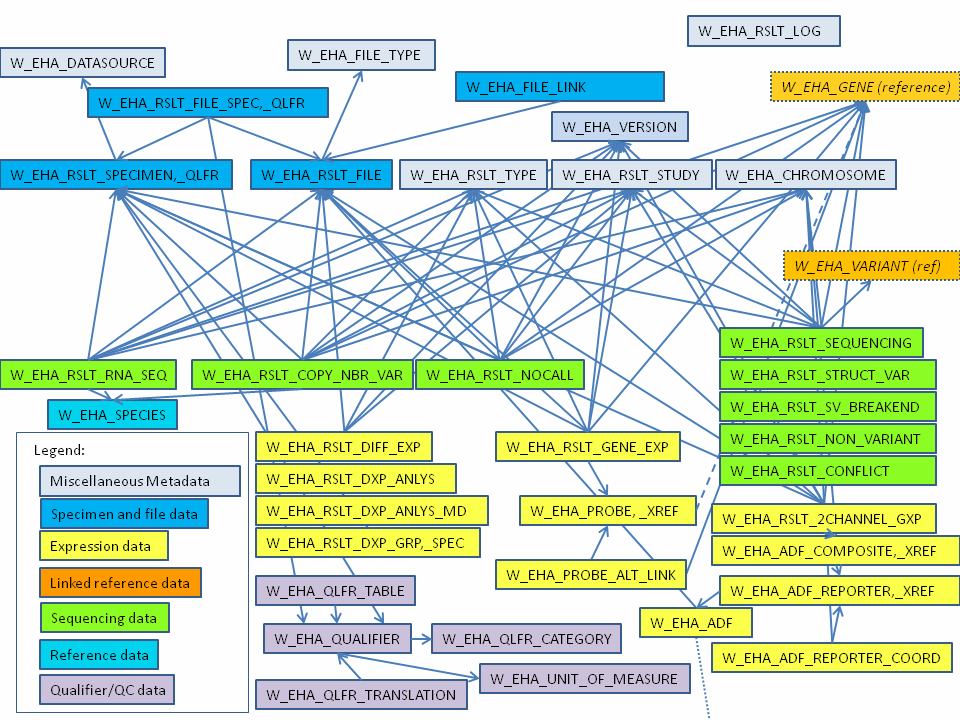
Figure 1-2 shows how the result tables link to create the ODB result tables section. Only table names are shown in the figure.
Figure 1-2 Result Data Logical Model (Core Tables Only)
The reference data starts with the W_EHA_SPECIES and W_EHA_DNA_SOURCE tables.
The species table stores information about each genome in the database. The current model permits any number of species genomes to be loaded. You must specify species in queries if there are similar genes between the organisms being tested.
The table also stores the promoter offset value that is used to define the promoter region of each gene, linked to the species record, if no PROMOTER component is defined. If set, this value overrides the global promoter offset defined in W_EHA_PRODUCT_PROFILE table.
The W_EHA_DNA_SOURCE table stores multiple records for different reference DNA sequences for each species. Each cell in the species has a copy of this reference DNA. There are buffers of DNA considered to be the reference for each organism. These reference sequences are then used to map detected variations for each organism tested. The W_EHA_DNA_SOURCE table has a foreign key to W_EHA_SPECIES and a CLOB column to store the reference nucleotide sequence information. The DNA table uses a specific notation to keep track of each DNA base in addition to the four standard characters: A, T, C, G. There are additional characters used for sections that have not been sequenced (N) and other characters are used to represent other possible DNA bases. The records in this table are used as the parent records to map genes and gene components. If Ensembl releases patches that illustrate how some genes are re-defined, new W_EHA_DNA_SOURCE records are created and linked to the other records, as required. This table also stores the chromosome location, which is described later. The position of features such as variants, segments, and so on in ODB is 1-based, following the standard from Ensembl. With the introduction of multiple reference support, W_EHA_DNA_SOURCE table links each reference DNA Source record to its DNA version, which is stored in the W_EHA_VERSION table under version type 'DNA'.
Figure 1-3 Genome Division by the Data Model
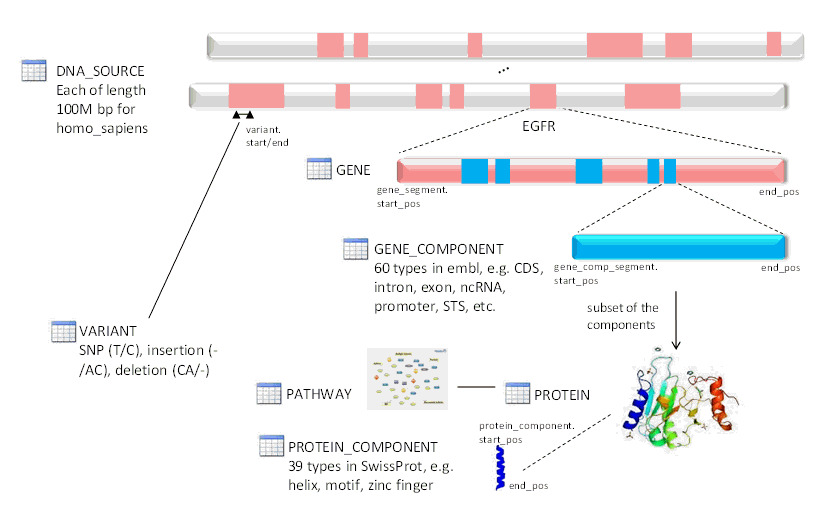
Each chromosome encodes for many different genes, each of which has a start and end position. The entire start-to-end region of the gene typically does not create the protein directly. Instead, there are recognized sections with the start-to-end region that scientists agree should be considered as part of the gene. The W_EHA_GENE table models how the Ensembl database refers to the gene, and the recognized gene name. The recognized gene name is maintained by HUGO Gene Nomenclature Committee (http://www.genenames.org/). This reference information is loaded into the model to provide accurate chromosome information for each gene as the patch DNA sequences loaded do not list chromosomes.
This table is required to map the different segments of a gene to DNA Source buffers. Some genes are sequenced in multiple buffers and require this joining table to track each segment. This table provides the location in the buffer and a sequence number to keep track of the order of each segment that composes the gene. There is also a COMPLEMENT field that is used to indicate if the coding strand of a gene is on the reverse strand (COMPLEMENT=1) or forward strand (COMPLEMENT=0) of a chromosome.
The _XREF suffixed tables associated with many of the different tables are used to list all the cross reference information stored in the Ensembl, SwissProt, and other databases. There is a finite list of databases used, and each database has a specific format for the reference ID. You can use these reference ID values for queries. The _QLFR suffixed tables associated with other tables are used to list other attributes. Each object can have an unlimited number of attributes such as /note that provides information to annotate the object. The database model stores this annotation data for reference.
The process of gene transcription is accomplished by many different interim molecules that originate from sections of the gene. For each protein created, there is a distinct set of sections which are used. Each of these groups is identified in the EMBL file by having the same TRANSCRIPT_ID qualifier. A GENE_STRUCTURE record is created to link to the protein and be used as a parent record for all of the gene components. A given gene can have multiple proteins that are created (sometimes using the same sections) and each has a different structure. Also, earlier research may have incorrect gene structures and the information is kept for historical reasons.
This table is used to store the various gene components. The EMBL file has many different objects listed (mRNA, CDS, STS, tRNA, misc RNA) and each has a specific meaning. Views are used to group the various types of objects in case there are queries to find genetic results that intersect with various gene regions. More user-friendly names are given (such as mRNA = MESSENGER_RNA, CDS = CODING_REGION, STS = STRUCTURAL_SEGMENTS, and so on). Various queries can be run searching for mutations that occur in any of these regions, including the entire gene region.
This table is used to link each component to W_EHA_DNA_SOURCE records. Many gene components have joined sections. Sometimes the joined sections are detected in different source buffers as well and the foreign key to DNA_SOURCE is required for each part of the gene components. There is a sequence number to keep track of the order of each section used in the gene component.
Most of the known genes produce different types of protein molecules. The EMBL files list the amino acids that comprise each protein molecule, and use an identifier for each protein molecule. The SwissProt files contain both amino acid (AA) sequences and additional information about the protein molecule. There are more descriptive names for each protein that are not stored in the EMBL files (such as, insulin). The W_EHA_PROTEIN table only stores AA sequences, and facilitates merging data from EMBL and SwissProt files for the proteins having the same sequences. This table is unversioned, and the additional data for proteins is stored in a separate table, W_EHA_PROT_INFO.
This table stores information about proteins, loaded from SwissProt files. It is linked to the W_EHA_PROTEIN table containing the AA sequences. Multiple W_EHA_PROT_COMPONENT records can be linked to a single W_EHA_PROT_INFO record. This table is versioned, that is, each record in it is associated with one or more versions of type 'PROTEIN' (through the W_EHA_PROT_INFO_VERSION table). This multi-version link is unique to the W_EHA_PROT_INFO table. It is used to prevent duplicates of the Protein information records (and dependent records in other tables) that have not changed from version to version.
This table stores all the protein components that are loaded from SwissProt files. This data may also be important for queries or reference. It can be important to show changes that may be occurring when variants are detected in the gene regions used to generate the amino acids of the protein.
The VARIANT table is used to record the known reference sequence, REFERENCE_SEQ, corresponding to one or more variants that differ from the reference DNA_SOURCE. Most of these variants are well documented and compiled from other research. When results are uploaded, sometimes novel variants are detected and there are no known references for this variant.
These results generate new VARIANT records, which may be of interest to researchers. There is a STATUS field which is used to indicate NOVEL or KNOWN variants. Since this table is queried frequently, it is quite large and requires partitioning. The VARIANT table has a foreign key to the DNA_SOURCE record, not the GENE record. This is because some genes may overlap, and there may also be several structures that are affected by a variant. This table is used to create result foreign keys as described later.
The Variant table contains a column PRECEDING_BASE, which stores the nucleotide base value preceding an insertion or deletion. The column, SVTYPE, stores the type of the structural variation, including insertions (ins), deletions (del), duplications (dup), copy number variations (cnv). The preceding base value is given, if and only if, SVTYPE value is not null for a variant record.
This table has a foreign key to W_EHA_VARIANT table and only stores the allele value for the large structural variant coming from the VCF file. This value comes from the ALT column present in the VCF file.
This table is very important to store reference seed information needed for identifying gene locations. The EMBL files report each gene with a LOCUS_TAG, which uses the registered name with the HUGO Gene Nomenclature Committee (http://www.genenames.org/). The entire reference data from this group is loaded as seed data in this table. The patch sequences (which are corrections to the human genome project) list the chromosome using the accession number of the DNA used for detection. The W_EHA_HUGO_INFO table is required to look up the HUGO gene names to find various cross references and the correct chromosome number for each gene. Each gene in the HUGO_INFO table is linked to a specific version of HUGO reference data.
Pathway is used to describe a series of interactions in a cell. Numerous biological pathways exist, including genetic, metabolic, signaling, and so on. This table is used to store publicly available pathways. Each pathway's participants are defined in the PATHWAY_PROTEIN table which has a foreign key to the PATHWAY table. Each pathway in this table is linked to the W_EHA_VERSION table with specific version of Pathway Commons build release.
This table is used to track which gene or protein belongs to a particular pathway. It has a foreign key to the PATHWAY table which associates a gene or protein with one or more pathways.
This table stores information relevant to SIFT (Sorting Intolerant From Tolerant) or Polymorphism Phenotyping (Polyphen) algorithms scores. SIFT or Polyphen are publicly available algorithms describing the impact of each variant on the resulting gene structure. The impact is evaluated both as a numeric score and a formal annotation, such as deleterious, probably damaging and so on.
Polyphen is an automated tool for predicting the possible impact of an amino acid substitution on the structure and function of a human protein. This prediction is based on straightforward empirical rules which are applied to the sequence, phylogenetic, and structural information characterizing the substitution. Possible annotation values are probably damaging, possibly damaging, benign, and unknown.
Sorting Intolerant From Tolerant (SIFT) predicts whether an amino acid substitution affects protein function. This prediction is based on the degree of conservation of amino acid residues in sequence alignments derived from closely related sequences, collected through PSI-BLAST. Possible annotations are tolerated, and, deleterious.
This table stores the possible annotations for SIFT or Polyphen algorithms that are mentioned above, such as probably damaging, possibly damaging, deleterious, tolerated, and so on.
This table stores variant impact or effect as computed by an Oracle proprietary script (stored procedure) based on variant effect on the resulting protein. The possible values of net effects are as follows:
Unknown — used when a variant is not in a coding region, or intronic, or crosses splice boundaries that would affect translation in unknown ways.
Frame-shift — used for insertion, deletion, indel variants that add or remove a number of nucleotides not divisible by 3 (the size of a codon).
Nonsynonymous - missense — signifies that a single nucleotide change results in a codon that codes for a different amino acid.
Nonsynonymous - nonsense — signifies that the stop codon occurs abnormally from the variant data in the coding region.
Synonymous — signifies that the variant in the coding region does not cause an amino acid change.
Note:
The variant impact script has been temporarily removed from ODB 3.0.This table stores default global Promoter and Flanking offsets which are the inputs provided during installation. It also stores various log level flags and DBMS output for data load ETLs and other procedure calls. Another column in this table 'VCF_FORMAT' stores a list of data types for the FORMAT column in the VCF file. Promoter offset is used during querying (through Promoter view: W_EHA_PROMOTER_V) and can be changed at any point. If PROMOTER_OFFSET in W_EHA_SPECIES is set, this promoter offset takes precedence over the one in W_EHA_PRODUCT_PROFILE.
Flanking offset should always be set to a value greater than Promoter offset. Promoters are assumed to fit within the Flanking offset region.
Changing the Flanking offset requires reloading all results tables which use genomic coordinates, such as sequencing and copy number variation result data. It is imperative to keep Flanking offset unchanged.
The logging level flags that can be set by the user include: Warning, and Info (which are set by default to 'Y'); Debug, TRACE, and DBMS output which are set by default to 'N').
This table lists the current version of the Omics Data Model and is used primarily by the Cohort Explorer application user interface (not included in this release). For example, currently it is set to 3.0.
This table stores names of diseases with possible genetic linkage.
This table stores literature-derived associations between diseases and genes that might contain disease causing mutations. It aggregates the mutation-disease linkage reported in the W_EHA_DISEASE_G_VARIANT table and associates the whole gene sequence with diseases caused by mutations in the gene. It also contains disease linkage for genes with disease causing variants for which no exact genomic coordinates were provided.
This table stores disease linkage for variants with known genomic coordinates. It includes linkage confidence provided by HGMD curators.
This table stores DrugBank-derived drug names.
This stores drug and gene associations linking DrugBank-derived drug names to their therapeutic targets.
This table stores the configuration information for the Array Data Format (ADF) files, which contain the annotation data required by Two-Channel gene expression result datasets. An ADF dataset loads into the W_EHA_ADF_* reference tables described below. Each file load creates an ADF record and the records inserted into W_EHA_ADF_COMPOSITE and W_EHA_ADF_REPORTER will have an 'ADF_WID' foreign key column to this table.
This table stores the gene composite elements associated with the 2-channel result data present in W_EHA_RSLT_2CHANNEL_GXP table. The composite element coordinates are input from the array design file (ADF) for the AgilentG4502A_07 platform. An additional table W_EHA_ADF_COMPOSITE_XREF is kept to store any external cross reference data for composite elements.
This table stores the probe (reporter) elements associated to the composite gene element present in W_EHA_ADF_COMPOSITE table. The reporter identifiers are input from the array design file (ADF) for the AgilentG4502A_07 platform. An additional table W_EHA_ADF_REPORTER_XREF is kept to store any external cross reference data for Reporters.
This table stores the probe (reporter) elements genomic coordinates associated with the reporter indentifiers present in the W_EHA_ADF_REPORTER table. The reporter genomic coordinates are input from the array design file (adf) for the AgilentG4502A_07 platform.
This table holds probe information for gene expression results, and each probe is designed to represent a particular gene. Since probe design varies by vendors, there may be multiple probes that correspond to the same gene. In the rare instance where more than one gene matches a probe, the model has a W_EHA_PROBE_ALT_LINK that should be manually populated. W_EHA_PROBE must be populated by the expression loader prior to loading any results corresponding to gene expression. In addition, any reference information pertaining to probes can be recorded in the W_EHA_PROBE_XREF table.
This table stores the link between gene names, somatic variants reported to COSMIC, both non-coding and coding, its sample annotation, the sample's reference keys to Histology and Anatomical site data, and mutation related annotations. The table has a reference key to W_EHA_VARIANT table as a main link to other reference source tables.
This table contains cross reference data to CANCER S(SUBJECT) VARIANT bridge table. The primary cross reference data stored in this table are PubMed publication IDs of associated somatic mutations.
This table contains cross Reference data to CANCER S(SUBJECT)_GENE bridge table. The primary cross reference data stored in this table are PubMed publication IDs of associated somatic mutations.
This table links gene annotation including Refseq and/or Genbank, and Ensembl gene IDs to somatic mutation cancer sample data submitted to Cosmic. The table stores a count of variants and linked gene mutations that is found for every gene symbol (hugo name) reported for each reference sample ID given by COSMIC. Each sample links to its histology and anatomical site references.
The table contains information on Reference samples studied for observed somatic mutations, which were submitted to COSMIC. The table provides sample annotation including cosmic tumor and sample IDs, sample source, tumor origin along with sample name. The table also references Histology and Anatomical sites associated to Samples.
This table stores the primary descriptive name of an anatomical site of a linked reference sample.
This table stores the descriptive name of an anatomical site subtype of a linked reference sample.
This table stores the primary Histology description of a linked reference sample.
This table stores the descriptive Histology subtype name of a linked reference sample.
This table stores the parent record to name a set of genetic codon translations to amino acids.
This table stores the link that each codon has with its corresponding Amino Acid.
This table maps how GEN_CODE tables are used with each species and chromosome.
The model currently supports the following two major categories of results:
Sequencing
Gene Expression
Types of sequencing results include simple variants, copy number variation, and no-call. Gene expression results comprise of regular microarray gene expression or RNA-seq results. Overall, results are populated into the following major tables:
W_EHA_RSLT_SEQUENCING
W_EHA_RSLT_GENE_EXP
W_EHA_RSLT_NOCALL
W_EHA_RSLT_RNA_SEQ
W_EHA_RSLT_2CHANNEL_GXP
W_EHA_RSLT_COPY_NBR_VAR
W_EHA_RSLT_NON_VARIANT
W_EHA_RSLT_CONFLICT
W_EHA_RSLT_SV_BREAKEND
All the major result tables listed above contain foreign keys to the following tables:
W_EHA_FILE_LOAD
W_EHA_RSLT_STUDY
W_EHA_RSLT_SPECIMEN
W_EHA_RSLT_TYPE
W_EHA_GENE
W_EHA_VERSION
Each record in these tables is linked to a specific reference 'DNA' source Version Label in W_EHA_VERSION table by the VERSION_WID foreign key. Additionally, where possible, each record is linked to a specific W_EHA_GENE record to allow for a gene based partitioning of these result tables. Those records that cannot be linked to a gene record have a GENE_WID value of '0'.
For Exadata, some of the result tables listed above have counterpart tables created dynamically during install, having the same columns as existing result tables. The new tables added are named using the same name as the parent result table with a _CHR suffix. These tables include:
W_EHA_RSLT_CONFLICT_CHR
W_EHA_RSLT_COPY_NBR_VAR_CHR
W_EHA_RSLT_NOCALL_CHR
W_EHA_RSLT_NON_VARIANT_CHR
W_EHA_RSLT_RNA_SEQ_CHR
W_EHA_RSLT_SEQUENCING_CHR
W_EHA_RSLT_SV_BREAKEND_CHR
For details on chromosome partitioned tables, see Section 7.11, "Chromosome Partitioned Tables".
This table contains sequencing results, more specifically variant information. It has a foreign key to the W_EHA_VARIANT table. The records in this table are linked to the record in the variant reference table. Information such as insertion, deletion, or substitution is recorded in this table along with any quality metrics on this information. The result sequencing table also stores large structural variants.
Note:
A record in the W_EHA_RSLT_SEQUENCING table may come from any of the four result file types, namely gVCF, VCF, MAF, or Complete Genomics masterVar file.This table contains results coming from VCF, gVCF and Complete Genomics sequencing files. Only CGI masterVar format records no-call results, that is, instances when there is incomplete information to make a call regarding variant information on an allele.
This table contains the non-variant information belonging to a specimen coming from VCF and gVCF files. The VCF loader populates this table when used in either 'GVCF' mode or 'NON-VAR' mode, provided the file contains non-variant information.
This table contains the low quality score variants which conflict with an existing high quality score variant reported in W_EHA_RSLT_SEQUENCING table for the same specimen. This table is populated through the VCF loader with data coming from gVCF files.
This table contains the structural rearrangement data coming from VCF files.
This table contains copy number variation results coming from the Complete Genomics platform and data from Affymetrix Genome-Wide Human SNP Array 6.0 along with any relevant quality or count metrics. This table is populated through the newly provided CNV loader, which can load the .SEG format file, and has been verified to load data from TCGA belonging to Affymetrix Genome-Wide Human SNP Array 6.0. No loader exists to load data from CGI format, however, the tables are designed to accommodate any attributes specific to CGI CNV data.
This is an additional table to store less frequently used sequencing metadata from the input file. It is always used as a helper table along with the main RSLT_COPY_NBR_VAR table.
This table is loaded from gene expression results and permits storing gene intensity measurements and quality metrics such as p-value and call information. A record can be inserted into this table only if the specified probe already exists in the W_EHA_PROBE table to establish a foreign key relationship.
This table is loaded from TCGA RNA SEQ file format for RPKM expression information of exons. The supplied loader only supports the loading of the exon version of the data files. TCGA has 3 different types of files: exon, gene, and splice junctions. Only the exon files are measured by exact chromosome locations. The other two file types are calculated estimations based upon gene locations using the exon data file.
This table is loaded from TCGA – Level 3, AgilentG4502A_07 platform specific, microarray dual channel gene expression files. For each gene record, the loader resolves two values it loads to the result table:
ADF_COMPOSITE_WID - by looking up the gene composite name in w_eha_adf_composite table.
GENE_WID - by looking up gene segment table for all complete and partial overlaps of gene segments in reference to gene composites genomic coordinates.
The Array Design file information associated with this data is loaded in the ADF Composite and Reporter tables in ODB, described in the Reference tables section of this chapter.
This table is a pre-seeded table with the types of results currently supported by the model. The result types are listed in Section A, "Additional Result Tables". Each record in the W_EHA_RSLT table supports a single specific result type.
This table permits each result to be linked to a study if specified by the end-user during loading. The study table is intended to be a shadow copy of a table in the clinical data model, called the Cohort Explorer Data Model study table, and only holds the study name and description. This table should be populated by the end-user before loading any results pertaining to a given study. The presence of this table enables partitioning the results into groups based on a study for which the results have been collected. Depending on the customer data and query requirements, by-study partitioning can be used as an alternative partitioning scheme to by-gene partitioning. Currently, partitioning by gene is the default.
This table holds all the chromosome names and is pre-seeded with names for all Human chromosomes. Refer to Section A, "Additional Result Tables" for pre-seeded data information. A result record in W_EHA_RSLT_SEQUENCING, W_EHA_RSLT_NOCALL, W_EHA_RSLT_COPY_NBR_VAR may be linked to a particular chromosome. As with the W_EHA_STUDY table, this table is used to partition results for improved query performance.
This table stores a list of aliases to chromosomes present in the W_EHA_CHROMOSOME table. Initially this table is seeded with a common chromosome alias list.
The W_EHA_RSLT_SPECIMEN table is linked to all result data tables. Every record in any of the result tables must have a foreign key that links to a particular specimen in this table. The W_EHA_RSLT_SPECIMEN table in turn, links to the W_EHA_DATASOURCE table which holds information about the database a given specimen comes from. This information, along with additional fields in SPECIMEN table such as SPECIMEN_NUMBER and SPECIMEN_VENDOR_NUMBER, can be used to uniquely identify and pull more metadata about a given specimen from other source systems. This information is not stored in the ODB. If the specimen database is CDM, two more tables are used: W_EHA_SPEC_PATIENT and W_EHA_SPEC_SUBJECT, linking a W_EHA_RSLT_SPECIMEN record with a CDM pationet/subject.
This table stores information regarding specimen sources. Each genomic result must have a specimen record connected to it coming from another schema with patient results. Specimen identifier is the link between the clinical results and genomic results. This table needs to be populated by the user prior to running any result loaders. If ODB is to be used with Cohort Explorer data model, this table should be seeded with one source of specimen samples, which is the Cohort Data Model.
This table is pre-seeded with file types supported by then loaders into the model. The list of valid pre-seeded values is specified in the appendix. W_EHA_FILE has a foreign key into this table.
These tables contain the header information from VCF and gVCF files. Any line starting with '##' in the VCF and gVCF file is stored in the File Load Qlfr table. This is also where the alternative file location is stored, if the user optionally chooses to specify -alt_file_loc in the argument list when loading results.
This table stores the foreign key to W_EHA_FILE and W_EHA_RSLT_SPECIMEN and basically links a specific file which is loaded to ODB by a loader to one or more specimen.
The ODB data model has new tables for Qualifier Metadata attributes. These tables include:
This table describes qualifiers - flexible attributes used in _QLFR tables. Qualifiers extend the concept of name/value pair attributes: they could be assigned to one of three data types - CHARACTER, NUMERIC and DATE and are grouped into functional categories. Units of measure can be specified for numeric qualifiers.
Qualifiers can be grouped based on functional categories. For example, a user might want to create qualifier categories such as DNA Sequencing, Gene Expression, RNA Sequencing, and CNV.
This table lists all qualifier tags applicable to a specific table.
This table holds names of a unit of measure.
This table stores translation rules to convert values from one unit of measure into another.
This is the fact table that stores key or value pairs for flexible attributes associated with a particular result file or specimen combination.
Each _QLFR table has a QLFR_WID attribute that points to a QUALIFIER_TAG record in the W_EHA_QUALIFIER table. It also has a foreign key attribute that references a record from a base table. For W_EHA_RSLT_FILE_SPEC_QLFR table the base table is W_EHA_RSLT_FILE_SPEC.
Three separate attributes are used to store character, numeric and date values. If a qualifier data type is NUMBER the QLFR_NUMB_VALUE attribute is populated, else it is empty. Similarly QLFR_DATE_VALUE field is populated for the DATE data type qualifiers. The QLFR_CHAR_VALUE attribute holds a reported value and is populated for any qualifier data type.
Newly enhanced, the ODB data model hosts tables for differential gene expression analysis results. However, there is no loader provided to populate result set to these tables. These tables include:
The main table used to store results from differential expression files.
Stores a listing of differential analysis result-sets loaded.
Stores metadata for differential expression analysis result-sets.
Stores and describes a list of differential expression groups of specimens.
Links specimens to differential expression groups.
All loaders and procedures created in the ODB output will log records to a single table.
This table stores logging information from all loaders and jobs. The column RESULT_ TYPE_NAME specifies the loader populating a given logging record. Each record stores species, specimen, datasource, OS user, host, and etl_proc_wid details taken from a loader run. The LOG_LEVEL column stores the logging level pertaining to a specific record. The log record includes details of record that caused a log entry, error info or trace fields, and a log summary field.
In queries for Gene Expression (single-channel) intensities are often compared not to a set value, but to the minimum (maximum, average, and so on) of all intensities for a hybridization or a probe. The queries, performing aggregation, are rather inefficient. To make them more efficient, the following two aggregate tables have been added for Gene Expression:
W_EHA_RSLT_GXP_HYBRID_AGG
W_EHA_RSLT_GXP_PROBE_AGG
The first table aggregates over a hybridization, while the second aggregates over a probe. They have almost the same set of aggregated columns, such as MEDIAN_INTENSITY, AVG_INTENSITY, MIN_INTENSITY, MAX_INTENSITY, and so on. Each of them also has columns, by which aggregation is done on the W_EHA_RSLT_GENE_EXP table (FILE_LOAD_WID and PROBE_WID in W_EHA_RSLT_GXP_PROBE_AGG, and FILE_LOAD_WID and HYBRIDIZATION_NAME for W_EHA_RSLT_GXP_HYBRID_AGG).
These tables are updated automatically by the Single Channel Results Loader every time a new Single-Channel Gene Expression file load is performed, and are automatically used by the TRC UI 3.0, when building Gene Expression queries.
All file and file load tables are populated by the result loaders, the gvf and adf reference loaders where there is a requirement to store file metadata references for records inserted.
This table stores the name of the input file used to load a result dataset. The file contains, along with the file name used, a unique global identifier (URI) for the file. This identifier is generated, if not provided by the user, on result load.
This table stores a record for each file load for a file. The table contains the field file_load_seg_numb that counts the number of times the file with the same URI is loaded into ODB. The table is referenced by all main result data tables and the view W_EHA_FILE_LAST_LOAD_V that displays the last file load record for a file with the same FILE_URI value.
The table is used to store metadata information for each file load.