6 Genomic Query
This chapter contains the following topics:
Genomic Query Tab
The Genomic Query tab can be utilized as a search interface into the patient genomic results in Oracle Healthcare Omics. Two search modes are provided.
Using Gene Search
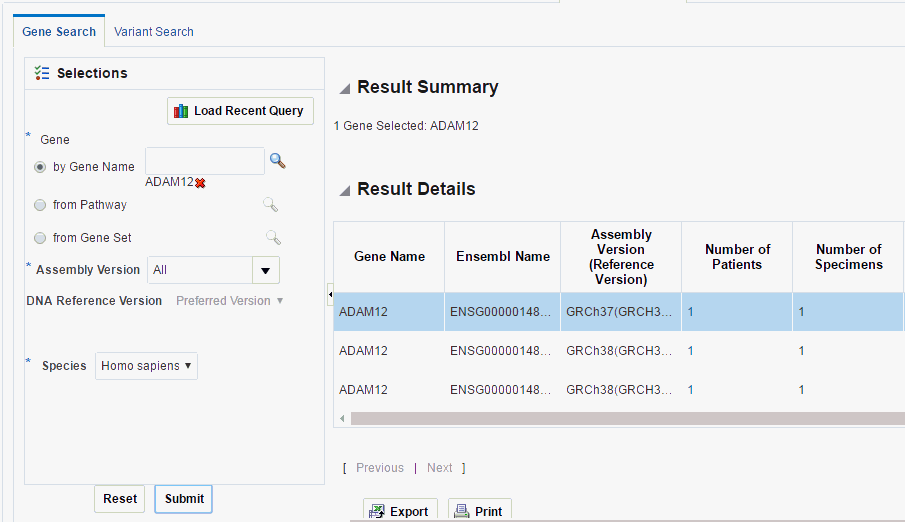
This tab lets you to specify one or more genes to be searched across results in OHO. To perform a gene search:
-
Navigate to Genomic Query > Gene Search.
-
To load a recently used query, click Load Recent Query. You can also search for the gene by gene name, pathway and gene set.
-
Enter the Assembly Version, Species and DNA Reference version.
Only the annotations of the selected genes are retrieved based on the DNA reference version. For example, if you select Assembly Version as GRCh37, then the DNA Reference Versions associated with GRCh37 assembly is displayed. The default is the one that is set as Preferred Version in Oracle Healthcare Omics (OHO, formerly known as ODB). You can also select another DNA Reference Version.The DNA Reference Version facilitates retrieving information such as Gene Name in the report.
-
Click Submit.
The results are displayed on the right. It includes the following:
-
Result Summary - this displays how many genes were searched for, how many have results found and how many do not have any results present in OHO for the criteria selected, what types of results are found if any.
-
Result Details - Tabular list of metadata for each gene that is found to have genomic results in OHO. The columns include:
-
HUGO gene name,
-
Ensembl gene name,
-
Assembly Version (Reference Version),
-
the number of distinct patients (or subjects) found with any data for a given gene,
-
the number of distinct specimen found with any data for a given gene,
-
the number of matching result files present in OHO
-
the type of result files present in OHO. For example, VCF, MAF, gene expression.
-
-
-
To export the resulting table to a spreadsheet, click Export.
-
To print the results, click Print.
-
Click the Patient count to navigate to different screens like View Record, Circular Genomic Viewer, Cohort List, Cohort Timelines, Cohort Reports and Genomic Data export.
If the pathway, geneSet or genes have been selected from the ad hoc genes selection, the result details table will be paginated based on gene selection. The first page displays the result for 25 gene selection and the next page will be the result for next 25 genes and so on.
Using Variant Search
This tab lets you to specify one or more variant identifiers to be searched across results in Oracle Healthcare Omics (OHO, formerly known as ODB). To perform a variant search:
-
Navigate to Genomic Query > Variant Search.
-
To load a recently used query, click Load Recent Query. You can also search for the variant by variant ID or attributes.
You can specify Genomic Position by typing in chr#:from-to location. For example, to find variants in base pair region 1-200 on chromosome 7, you would need to type in chr7:1-200. Also, if you want to search for any variants on chromosome 7, you can enter chr7.
Additional options that can be specified include known or novel status on the variant, variant type such as insertion, deletion, substitutions, indel, complex. The strand is the directionality of the variant and it can be either plus (+) or minus (-).
-
Enter the Species and Assembly Version.
-
Click Submit.
The results are displayed on the right. It includes the following:
-
Result Summary - this displays how many variants were searched for, how many have results found and, if applicable, how many do not have any results present in OHO for the criteria selected, what types of result files are found for the variants if any.
-
Result Detail - Tabular list of metadata for each variant that is found to have genomic results in OHO. The columns list includes the following option:
-
Variant Name, if the name exists,
-
Location of the variant,
-
Hugo Name
-
Assembly Version (Reference Version)
-
Status, whether the variant is known or novel,
-
the replace tag that tells the end user what exactly the variant is,
-
Tag, whether this variant is an insertion, deletion and so on,
-
Impact, the impact of a given variant (only available for known variants),
-
Study, the study this variant has been found to have results for,
-
Number of Patients, the number of distinct patients found with any data for the particular variant,
-
Number of Specimen, the number of distinct specimen found with any data for the particular variant
-
Number of Result File, the count of type of result files present in OHO,
-
Result File Type, type of result files present in OHO, for example, VCF, MAF, gene expression with data for a given variant.
Note:
On gene search selections: A variant may be present in alternate genes that overlap with the gene region of the selected gene in a given version. For any variant found, these alternate gene annotations are displayed to the user along with annotations from gene of interest. -
-
-
To export the data to a spreadsheet, click Export.
-
To print the current screen, click Print.