| Oracle® Healthcare Precision Medicine User's Guide Release 1.0 E75998-01 |
|
|
PDF · Mobi · ePub |
| Oracle® Healthcare Precision Medicine User's Guide Release 1.0 E75998-01 |
|
|
PDF · Mobi · ePub |
This section describes all the tasks related to annotating variants. It includes the following sections:
To annotate variants, perform the following steps:
Open the report in the reporting view.
Select the variants you want to annotate.
Click Annotate on the top right. The Annotation Pane appears.
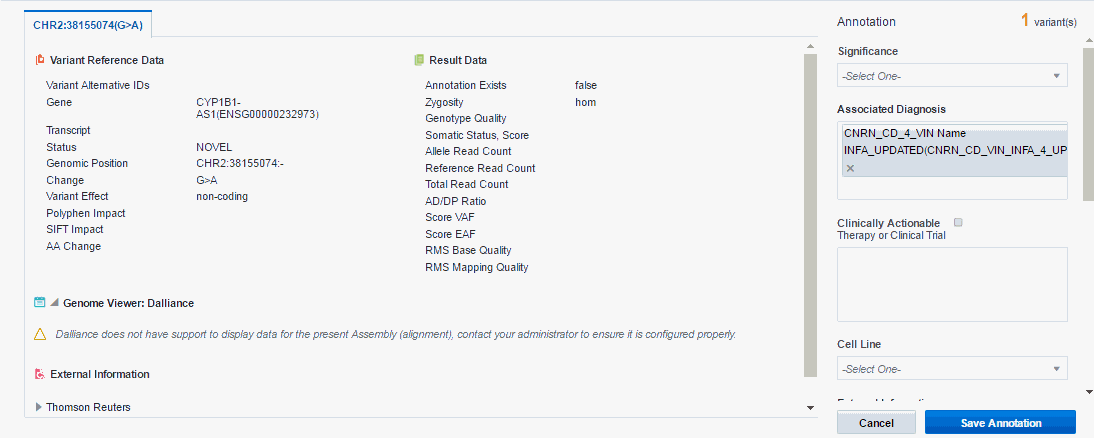
Enter details in the annotation pane.
Click Save Annotation. You will receive a confirmation.
Click Yes. The variant is annotated and the following icon appears next to it in the Reporting View.
The following entry appears in the Report Review pane.
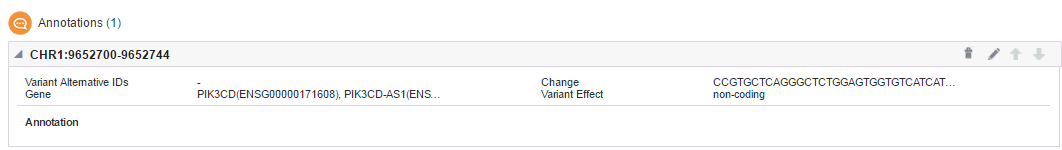
You can delete or edit this information using the icons on the top right of this section.

This view displays:
important reference details and metadata about each variant
details about the given variant based on the result file it appeared in
Only molecular pathologists and researchers have access to this feature.
Note:
A very small subset of variants may appear in multiple UI screens and reports to be associated with the placeholder record in the gene table with the name NO GENE.A NO GENE gene is not a regular coding gene and can be safely ignored when displayed, with any mapped variant assumed to be intergenic unless mapped to another valid gene region.
It lists the following information:
Variant Reference Data
Variant Alternative IDs - Displays any other names that the variant may have
Gene - The gene containing the variant
Transcript - The transcript being used to determine the impact
Status - Can be either known or novel
Position - The position of the variant within the chromosome
Change - Basepair change
Effect - Based on the preferred transcript, it is the effect the variant has on resulting protein. Among the effects are missense, nonsense, noncoding, frameshit and so on.
Polyphen Impact - The PolyPhen Impact of the variant
SIFT Impact - The SIFT impact of the variant
AA Change - Amino acid change
Result Data
Annotation exists - Indicates whether variant is already annotated in the report
Zygosity - Variant zygosity. Similarity of alleles in an organism to reference.
Genotype Quality - Genotype Quality
Somatic Status, Score - Calculated ration of somatic likelihood
Allele Read Count - Number of reads that support the given allele
Reference Read Count - Number of reads that support the reference sequence
Total Read Count - Total number of reads
AD/DP Ratio
Score VAF - Positive or negative integer representing confidence in the call from CGI masterVar file
Score EAF - Positive integer representing confidence in the call from CGI masterVar file
RMS Base Quality - RMS base quality at this position
RMS Mapping Quality - RMS mapping quality at this position
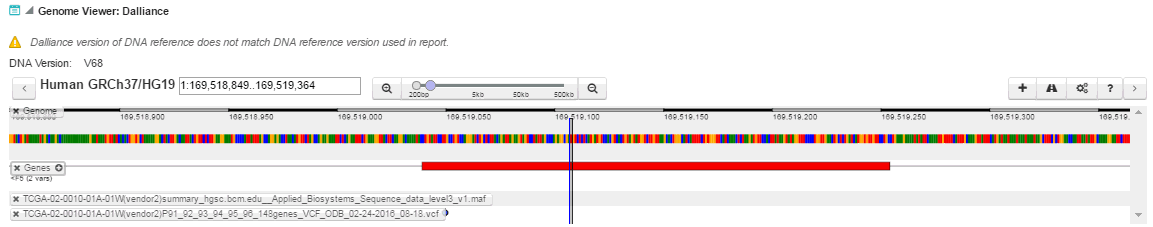
Genome Viewer: Dalliance - view variant and gene in embedded genomic browser screen against the annotation and reference or assembly information. The system checks if the Dalliance version of DNA display matches what is actively used for the given report, that is, the Preferred Version of DNA used for a given Assembly set.
External Information:
Thomson Reuters - If subscribed, displays the reference information for a selected variant. To view the view Thomson Reuters reference information, select the Variant and Disease from the respective drop-down lists.
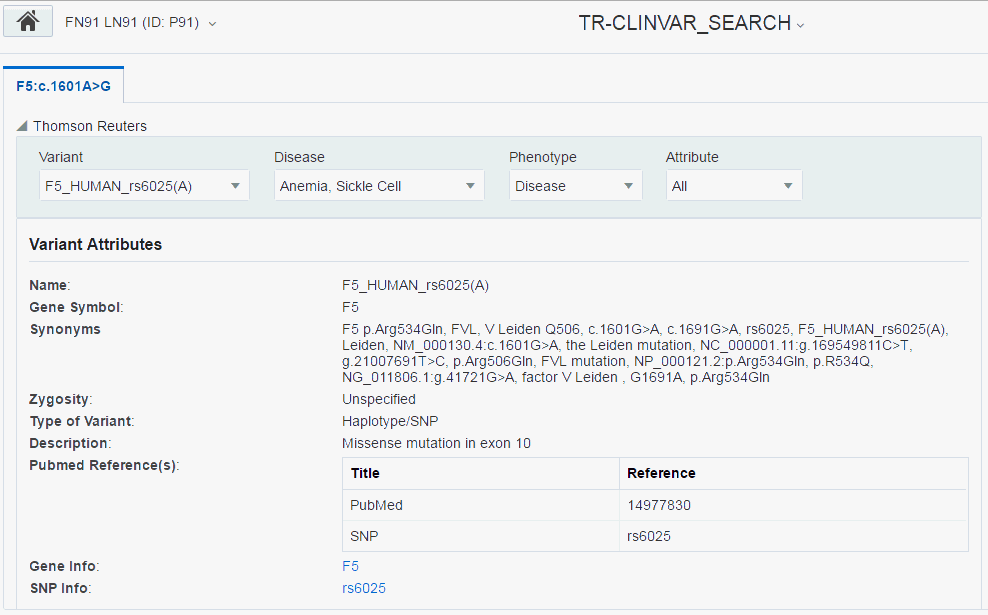
The following sections are displayed:
Variant Attributes
Disease Attributes
Casual Associations
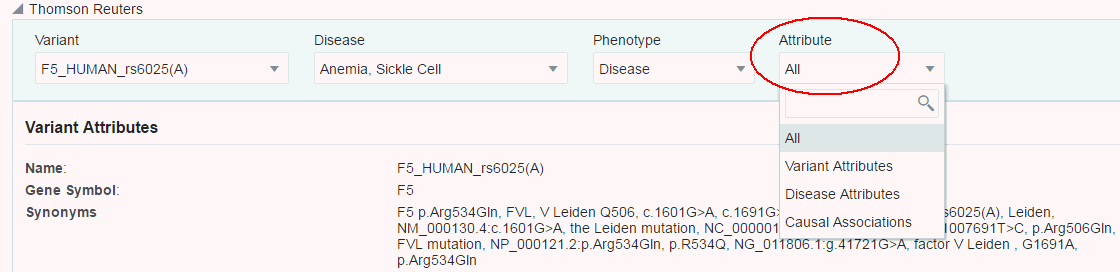
You can select which sections you want to view using the Attributes drop-down list. By default, all sections are displayed.
ClinVar - If subscribed, displays the reference information for a selected variant. Select a variant from the Variant drop-down list to view ClinVar reference information.
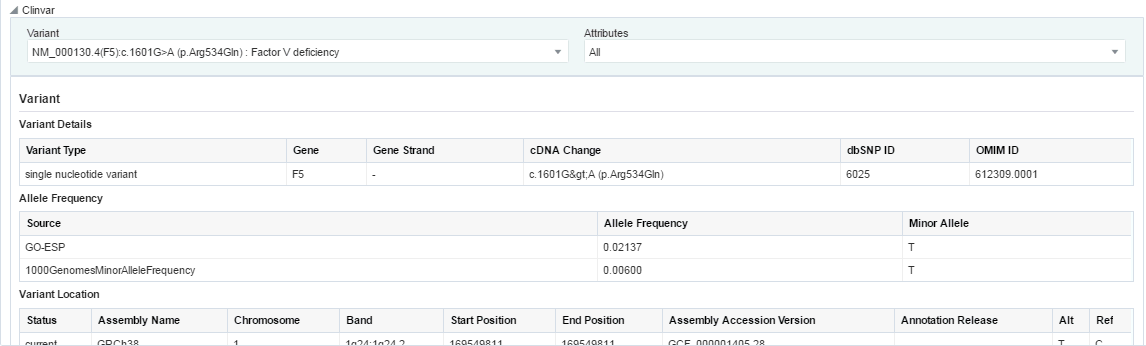
The following sections are displayed:
Variant - displays Variant Attributes including Variant Details, Allele Frequency, Variant Location
Disease Phenotype
Clinical Significance
ClinVar Submission Details
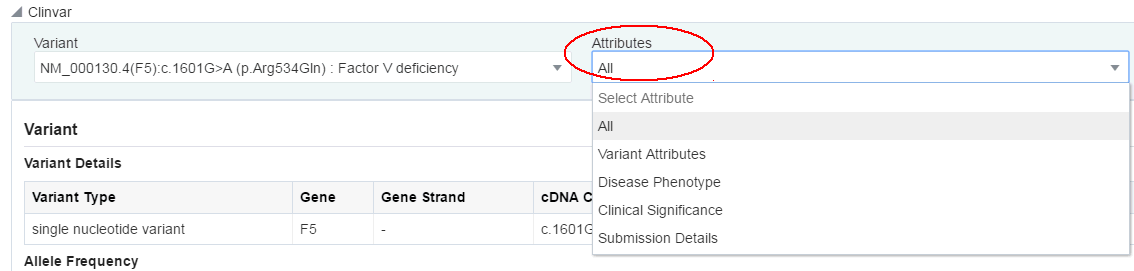
You can select which sections you want to view using the Attributes drop-down list. By default, all sections are displayed.
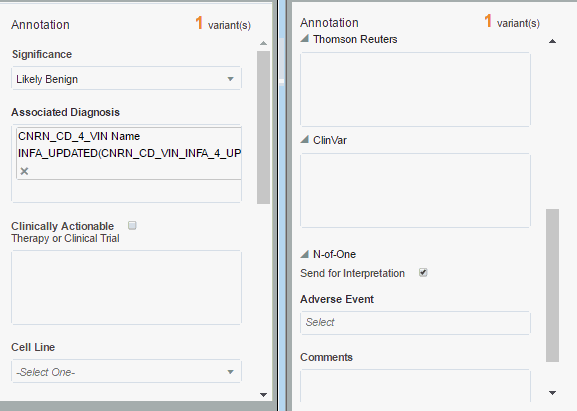
The Annotation Pane lets you annotate variants with your comments, functional labels and so on. This pane contains the following default fields and can be fully customized by the administrator:
Significance - has the values Pathogenic, Likely Pathogenic, Benign, Likely Benign, Unknown Significance. You can select any one or leave this blank.
Associated Diagnosis - diagnosis that is associated with the variant.
Clinically Actionable - this marks a variant as clinically important and highlights at the top of the report
Therapy or Clinical Trial - Enter any clinical trials or therapies that are currently available
Cell Line - has the values Somatic, Germline, Unknown. You can select one of them.
Annotation for Thomson Reuters, ClinVar - you can select relevant information from Thomson Reuters and ClinVar in the Tabular View and add this information to the annotations.
Note:
You must subscribe to Thomson Reuters and ClinVar to access their reference information.N-of-One - Selecting the check box Send for Interpretation will mark this variant as a candidate to send for N-of-One interpretation.
Adverse Events - select adverse events, if any, for example, Severe Allergic Reaction.
Comments - enter your comments on the report